True 3D spatial transcriptomics is transforming how scientists understand tissue biology and molecular activity
In this guest editorial, Jeremy Lambert shares how Stellaromics delivers true, high-resolution 3D spatial transcriptomics
19 Oct 2025
Jeremy Lambert, Director of Product Management, Stellaromics
“At Stellaromics, our vision is to unlock the next dimension of spatial biology by enabling scientists to study intact thick tissue structures in 3D with true subcellular resolution and multiomic readouts,” says Jeremy Lambert, Director of Product Management.
Stellaromics’ Pyxa™ is the first and only instrument on the market capable of delivering true, high-resolution three-dimensional spatial transcriptomics. The ability to uncover cell–cell interactions, tissue morphology, and molecular gradients in intact, thick tissue sections is already driving new insights into disease mechanisms and therapeutic development.
Pyxa is purpose-built for high-plex, 3D spatial transcriptomics in intact thick tissue. The system is built on STARmap chemistry, a published method for detecting transcripts at subceullular resolution in intact tissue up to 100 microns thick. To bring this technology to the world, Pyxa delivers a complete end-to-end process for sample preparation, automated imaging, and data analysis: streamlined reagent kits for simplified sample preparation, automated high-speed imaging for rapid data capture, integrated computational analysis, and intuitive 3D visualization tools.

The Pyxa™ platform offers sample preparation, automated imaging, and 3D data analysis all in one.
Pyxa enables researchers to profile both gene expression and translational activity at subcellular resolution across large tissue volumes. This sets Pyxa apart from other spatial transcriptomics platforms that rely on reconstructing 3D models from serial thin tissue sections which can distort delicate structures, lose cellular layers, and impose a high computational and labor burden on labs. Large cells such as neurons and structures such as blood and lymphatic vessels are not fully captured in thin tissue sections, nor is tissue anatomy such as cortical layers in the brain or the internal structure of the kidney. Instead, Pyxa captures biology in situ, maintaining tissue architecture and cellular context. “This is a leap forward in our ability to understand how cells interact in their native tissue context. We know that biological systems don’t operate in two dimensions, but now it’s possible to visualize and measure 3D interactions in situ,” says Lambert.
Importantly, 3D spatial transcriptomics on Pyxa is complementary to existing 2D platforms. Thin-tissue spatial transcriptomics approaches map broad tissue landscapes and remain useful for whole-transcriptome discovery or for analyzing very large tissue areas at lower resolution. Pyxa builds on those insights by allowing researchers to explore areas of interest in 3D, with greater resolution, depth, and multiomic context that 2D methods simply cannot provide.
Early adopters are using Pyxa to answer questions that were previously impossible to ask in fields including neuroscience, immunology, oncology, and developmental biology. “It’s motivating to see scientists around the world adopting Pyxa to uncover novel insights enabled with 3D analysis” says Lambert. “At Stellaromics, we have always believed that 3D transcriptomics would power novel insights into the molecular mechanisms behind disease, and it’s wonderful to see that coming to fruition with early adopters of Pyxa.”
One compelling example comes from the study of neurodegenerative diseases: using Pyxa, scientists have been able to visualize the clustering of immune and endothelial cells around plaques in the brain, indicating the progression of Alzheimer's disease. In immuno-oncology, scientists have used 3D spatial transcriptomics to detect rare, treatment-resistant cancer cells early in tumor evolution, before they become dominant. These studies reflect a transformative moment in our understanding of disease progression and therapeutic development, leveraging a true three-dimensional understanding of cell and tissue biology.
As the spatial omics field continues to evolve, Pyxa will be a foundational tool at the intersection of spatial biology, drug discovery, and clinical research. “We believe that the ability to visualize high-resolution, 3D spatial transcriptomic data will transform how we study biological drivers of complex diseases such as cancer, autoimmune diseases, and neurodegeneration,” adds Lambert. “Pyxa can help researchers pinpoint therapeutic targets, map cellular responses, and understand how interventions influence disease mechanisms with unprecedented precision.”
Pyxa enables researchers to gain novel insights into drug targeting and efficacy, for example in the context of cell and gene therapy, allowing scientists to visualize the precise localization of a drug and understand its effect on local gene expression with subcellular localization. In clinical research, this detailed, contextual information will be key to identifying novel biomarkers and understanding why some patients respond to a drug while others do not. The ability to create comprehensive 3D "disease atlases" of patient samples will empower scientists to move from a generalized view of disease to a patient-specific understanding of pathology. Stellaromics’ technology is poised to become an essential component of clinical trials and diagnostic workflows, providing the critical spatial insights needed to unlock the next generation of precision medicines.
Stellaromics offers early access to Pyxa through its Technology Access Services, allowing researchers to submit tissue samples to Stellaromics’ Boston laboratory and partner with its team to run pilot studies. The standard service offering is a 250-gene mouse neural cell typing panel for fresh frozen moue brain tissue. Custom panel design is available by request for the study of other mouse and human tissue types; recent tissue types processed through Stellaromics’ Technology Access Services include mouse kidney, mouse cerebellum, mouse retina, human liver, and human colorectal cancer. Data delivery includes data files that are compatible with numerous open-source tools for quantitative analysis, as well as access to PyxaStudio, Stellaromics’ proprietary software for visualizing 3D transcriptomics data.
“Our Technology Access Services customers have demonstrated the growing need for 3D spatial transcriptomics in a variety of fields,” explains Lambert. “Pyxa is enabling scientists to identify complex tissue structures, cellular neighborhoods, and rare cell types in their native context for the first time. This deeper view of tissue organization and molecular activity is revealing critical drivers of disease and accelerating our understanding of how to intervene effectively.”
Three-dimensional spatial transcriptomics is just the beginning of what Stellaromics believes is the future of spatial biology and its impact on biomedical research and drug discovery. On the horizon is the launch of the RIBOmap assay, which detects actively translated, ribosome-bound mRNAs – what is termed “translatomics.” Looking ahead, Stellaromics envisions more meaningful integration of transcriptomic data with other omics layers including epigenomics, proteomics, and metabolomics. Together, these technologies provide a unified view of gene and protein expression in 3D context, giving scientists a more comprehensive view of biological systems than ever before.
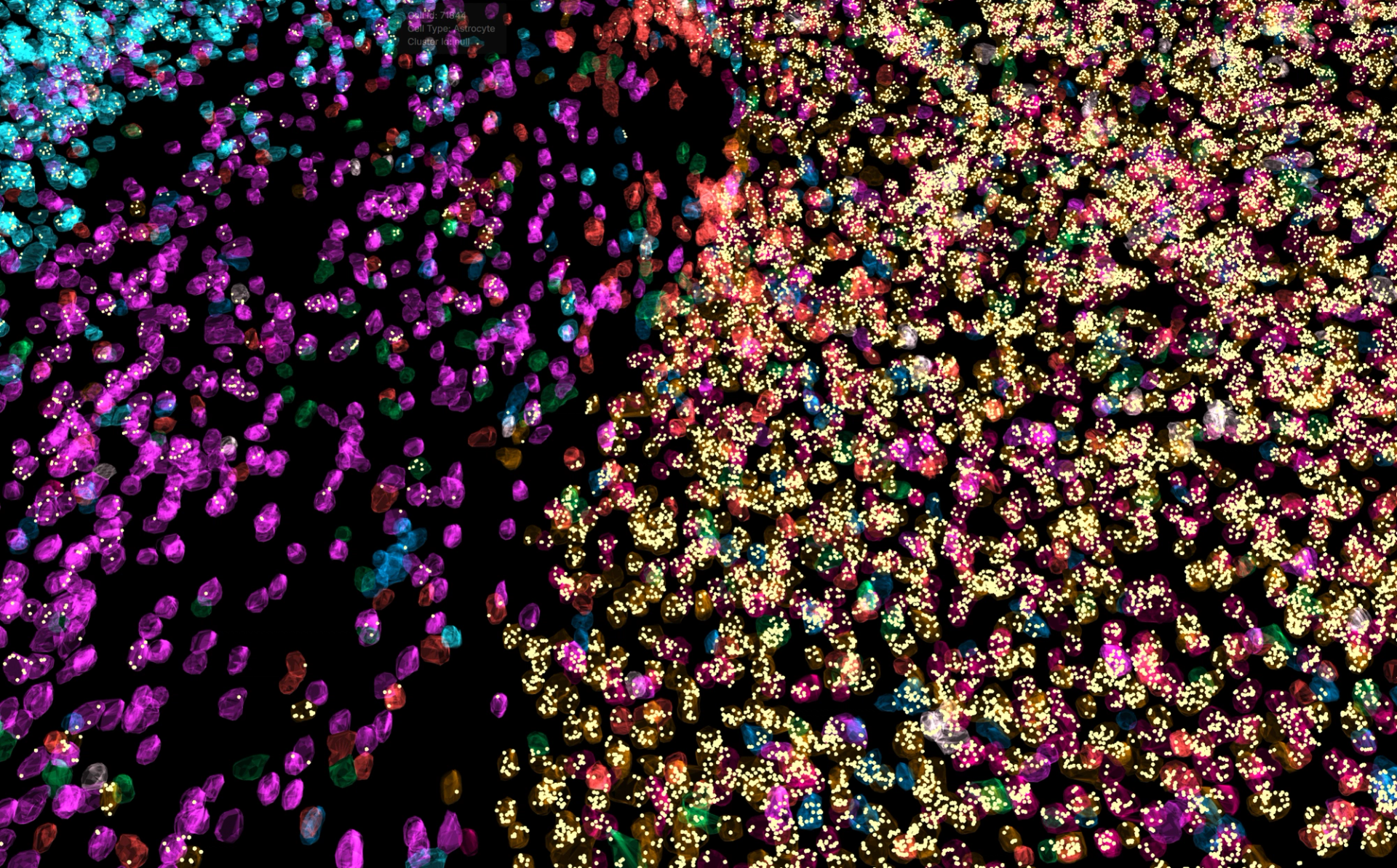
PyxaStudio software was developed to visualize cells and transcripts in three dimensions
