A 3-Dimensional ‘Google Earth’ Style Map for Tumors
Dr. Josephine Bunch, UK Grand Challenge awardee, describes her unprecedented research into a tumor’s intricate architecture and shares her ultimate goal of mapping what’s inside a tumor
22 Oct 2017
To fight cancer, scientists strive to know everything they can about it, so they can formulate an approach that makes it difficult for tumors to thrive. One of the seven grand cancer challenges identified by Cancer Research UK is to map tumors at a molecular and cellular level. Although cancer and its manifestations have been thoroughly studied, the unique components of a tumor remain unknown. SelectScience® speaks with UK Grand Challenge awardee, Dr. Josephine Bunch, at the National Physical Laboratory, London, to learn about her efforts in mapping tumors from every possible angle.
Dr. Bunch’s project is similar to creating a Google Earth style of maps for a tumor’s tiny microenvironment. For instance, to entirely map out a city, one would need to learn of its different neighborhoods (the ‘what’) as well as carefully determine their exact location (the ‘where’). Dr. Bunch plans on doing exactly that – finding out what molecules make up a tumor, and what positions they occupy within a tumor mass. In this video, Dr. Bunch highlights the ‘how’ of this momentous project i.e. the methods that she plans to employ to create this comprehensive tumor layout.
What’s in a tumor, and where is it?
“Our project has brought together a whole range of mass spectrometry techniques, along with other spectroscopy instruments like MRI, with a view to mapping as many constituents of the tumor as we can. By that I mean as many different kinds of molecules – small molecules and large molecules – over a whole new range of different scales,” says Dr. Bunch. “We want to be able to see molecules inside single cells, but also be able to map whole tissues and even make measurements during surgical procedures.”
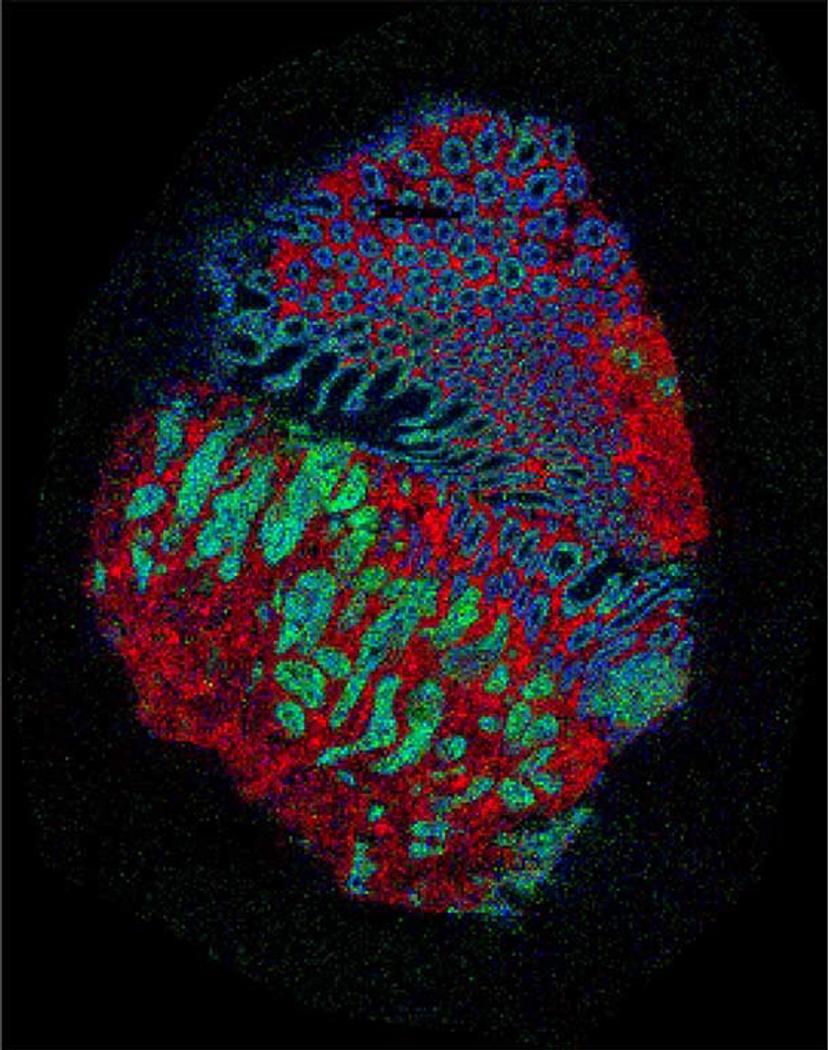
A picture of a bowel cancer sample made using mass spectrometry imaging. The artificial colors represent measurements of molecules and show different types of tissue – red is tissue that lines the bowel, green is the tumor and blue is muscle. Credit: Zoltan Takats, Renata Filipe-Soares (Imperial College London); Nicole Strittmatter, Gregory Hamm, Richard Goodwin (Astra Zeneca); Rory Steven, Adam Taylor, Alan Race, Spencer Thomas, Rasmus Havelund, Josephine Bunch (NPL).
All hands on deck to dive into cancer’s unknown territory
Dr. Bunch plans to use an exploratory approach to create these tumor maps using multiple spectrometry methods in tandem. This application of an analytical tool such as mass spectrometry towards deciphering tumor biology is unprecedented in cancer biology, which has relied heavily on molecular and fluorescent methods. The latter, however, works only if cellular and molecular targets are well-known.
“Many methods can produce really detailed information about the location of different molecules in a cell or a tissue. But typically, most of these methods require us to know what we’re looking for before we perform the experiment,” notes Dr. Bunch. “For example, optical microscopy will give us fantastic images of the distribution of different proteins but we have to know which protein we want to look for first. And usually, that would involve using an antibody conjugated with a fluorescent marker. That’s a very powerful method of looking for something that we already know,” she adds, commenting on the conventional approach in cancer research so far.
Dr. Bunch’s projects, however, will delve into the unknown. “What we want to do in this project is measure many things; we don’t know what we’re looking for yet. We’re trying to produce an unbiased review of the tissue biochemistry in a tumor,” she says.
“When we put together our project we realized that no single mass spectrometry imaging technique can address everything that we want to measure,” says Dr. Bunch. Several spectrometry methods will come together as she sets out to study tumor metabolism at different molecular levels. “We have a whole range of instruments available to us, starting from secondary ion mass spectrometry through desorption electrospray ionization (DESI), matrix-assisted laser desorption Ionization (MALDI) all the way through to mass cytometry and CY-TOF approaches,” she notes. Each of these techniques is useful for measuring different classes of molecules. “We can't really decide before we embark upon the project which of those are more important. So, we're bringing them all to bear on the same problem.”
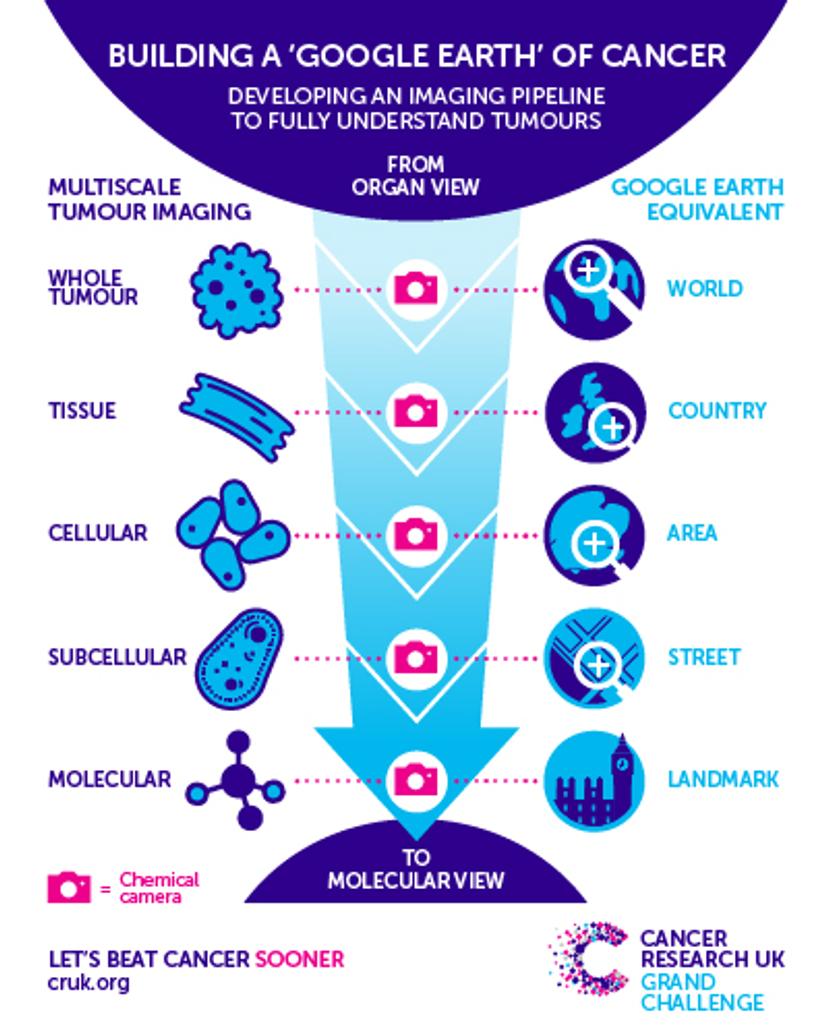
The top-down approach with which Dr. Bunch will map out a tumor involved multiple techniques used at tandem. Image courtesy of Cancer Research UK.
Pilot studies at the Bunch lab, in collaboration with other faculty members, include understanding the performance capacity of each of these methods to decide the spatial resolution a particular method offers. “We’re always balancing sensitivity with spatial resolution in mass spectrometry imaging methods,” says Dr. Bunch. “At the National Physical Laboratory, we have a range of MALDI and DESI instruments. The ones that we are using most routinely in the Grand challenge UK project at the moment are the Waters MALDI and DESI platforms… their SYNAPT instruments.”
The team works with colorectal tumor samples as well as samples from the pancreas, breast, and brain. They will employ a top-down approach, performing large high-throughput analyses first, and then honing into fine details of a tumor’s architecture. “We will be taking samples in sections using one of the methods, such as MALDI or DESI, which are our higher throughput methods to obtain our initial maps. We’ll then use a range of multivariate analysis tools and segmentation methods to find areas of particular interest in this tumor…probably several different areas of a single patient’s tumor,” says Dr. Bunch.
The impact of the team’s project will be extensive. Dr. Bunch adds: “I think by using these techniques in combination we're certainly extracting more information from a tumor sample than we’ve ever done before. By measuring the full scope of different molecules – small molecules all the way up to proteins at a range of scales, we are really able to review regions of interest in individual tumors in far more detail than ever before.”
One of the ultimate goals of the project is that the molecular data generated leads us to new ways of measuring tumors, in real time, in surgery or using non-invasive methods. Dr. Bunch concludes: “From a treatment point of view, the more we understand the molecular basis of tumors, particularly in a metabolic point of view, the better that we can design therapies, either new drugs or new intelligent surgical intervention.”
