Absolute Quantification of Protein and RNA in a Single Cell Using Digital PCR Technology
Read About the Pioneering Technique Published in Molecular Cell
1 Mar 2017
Dr Cem Albayrak, Assistant Professor of Chemical and Biological Engineering, College of Engineering, Koç University, spoke to SelectScience® about his research, which developed a powerful, novel methodology for the absolute quantification of messenger RNA (mRNA) and protein expression in single cells, using droplet digital PCR (ddPCR).
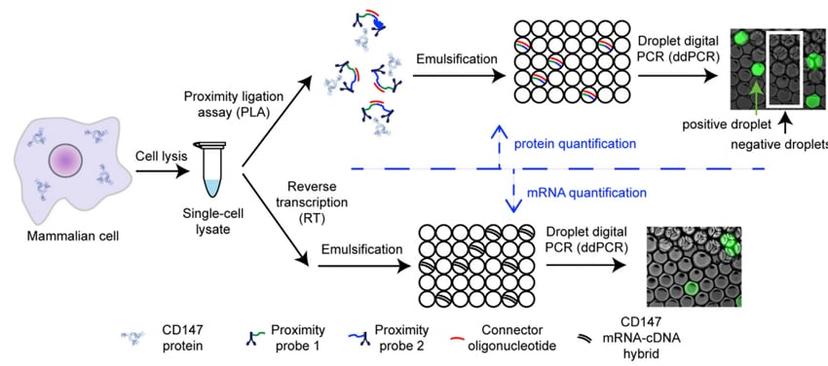
The Digital PLA Procedure schematic (Courtesy of Dr Albayrak).
The method was recently published by Dr Albayrak and colleagues in Molecular Cell (DOI: 10.1016/j.molcel.2016.02.030). Dr Albayrak completed the work as a postdoctoral researcher at the Department of Biosystems Science and Engineering (D-BSSE) at ETH Zürich in Switzerland. The work focused on the use of microfluidics and biochemistry to accurately quantify protein and RNA from single cells. The team combined droplet digital PCR technology with proximity ligation assay (PLA) to facilitate absolute quantification of both mRNA and protein in a single cell. The method, known as digital PLA, used standard digital PCR procedures combined with PLA.
Advantages of combining the methodologies
PLA is an immunoassay that enables high resolution and sensitive detection of proteins in situ. Different epitopes on the protein of interest are recognized by primary antibodies that are conjugated to short DNA probe sequences. When two antibodies bind their epitopes on the same target protein, the probe sequences come in close proximity and interact with connector DNA oligonucleotides. The strands can then be amplified using digital PCR to facilitate detection of the protein of interest.
Droplet digital PCR is a technology that enables the detection and quantification of very low levels of DNA or RNA. A sample is partitioned into nano-liter sized droplets and target DNA amplification is performed within each droplet using specific primers and fluorescently labelled probes. Fluorescent droplets are scored as positive for the target sequence, whilst droplets lacking fluorescence are scored as negative, which provides an absolute quantification of target DNA within the sample. By applying the ddPCR principle to the protein-antibody-conjugates, precise protein quantification was made possible from a single cell. mRNA levels from the same cell were also quantified using ddPCR, as indicated in the digital PLA schematic above.
In the published study, Dr Albayrak and his colleagues demonstrated that digital PLA was extremely sensitive in the absolute quantification of CD147 protein and its corresponding mRNA. The power of this type of dataset could enable researchers to model transcription and translation mechanisms in a single cell, and would be sensitive enough to detect low concentration changes in response to changing conditions.
Future applications
The use of the new digital PLA methodology has significant implications for the future of diagnostics. The remarkable sensitivity of the technique will allow minuscule concentrations of biomarkers to be detected. In addition, it would be possible to look at biomarkers in real-time, as compared to current methodologies, which can analyze end-point measures. The technique could also be applied to primary cells taken from patients and has many possible applications in precision medicine. However, there are still some limitations. Detection of proteins is limited by the availability of suitable antibodies and the technique is not yet sufficiently developed to enable multiplexing.
Dr Albayrak and his colleagues have developed an accessible assay that can precisely quantify mRNA and protein in the same sample using readily available and inexpensive methodologies.
Access the full research article here to learn more about this cutting-edge technique. Find out more about ddPCR in this video.

