Advancing clinical research with high-throughput proteomics
In this guest editorial, learn why high-throughput proteomics will be an essential tool to advance personalized medicine
3 Jul 2022

In this guest editorial article by Roman Fischer, Associate Professor in Clinical Proteomics, Target Discovery Institute, University of Oxford, and Gary Kruppa, Vice President of Proteomics at Bruker Daltonics, learn about the future of high-throughput proteomics and its role in clinical research.
High-throughput proteomics will play an essential role in advancing drug discovery and development towards a personalized, patient-centric approach to medicine.
With the global personalized medicine market set to be valued at $796.8 billion by 2028, this trend relies on significant technological and methodological advancements. Proteomics has emerged as an extremely powerful tool for protein scientists, biologists and clinical researchers.
The evolution of clinical proteomics has been guided by the development of applications used for spatial characterization of proteomes – the sets of proteins expressed at a particular time within a biological structure such as a cell or an organ. The expansion of analytical capabilities using integrative omics approaches has led to a more comprehensive picture of molecular processes in human disease and increased the demand from clinical researchers to include proteomics analysis of samples. New methodologies must now be developed for the proteome characterization of clinical cohort samples at high throughput.
A novel approach to proteomics
Blood samples are difficult to analyze due to the high dynamic range of protein abundance; and the limited speed, sensitivity, and resolution of mass spectrometers, makes achieving coverage of complete proteomes a challenge. Ongoing developments in mass spectrometry (MS) now enable researchers to visualize a larger fraction of the proteome of a cell or organ, with the goal of comprehensively identifying all proteins and their associated biological activities. They can use MS technology to uncover critical ‘molecular windows’ within complex disease processes that help to drive the discovery of new therapies.
One of the key breakthroughs in separation technology is trapped ion mobility spectrometry (TIMS), a technique where ions are propelled through the TIMS tunnel by a gas flow. The ion’s motion is controlled by an electric field, preventing them from moving beyond a position defined by the ion’s mobility, which is determined by its collision cross section (CCS). The unique design allows researchers to measure the CCS values for all detected ions, which can be used to further increase the selectivity of the system and enable a more reliable quantification. The design also allows for ions to be accumulated in parallel in the front section, while ions in the rear section are sequentially released depending on their ion mobility. This parallel accumulation-serial fragmentation (PASEF) technology synchronizes the quadrupole isolation mass window with the elution time of specific peptide packages from the TIMS funnel to achieve >100 Hz sequencing speed without losing sensitivity or resolution. Its superior results reduce both sample amount required and the frequency of MS maintenance.
Enabling studies for clinical research
As the field of clinical proteomics research has evolved, there has been a large increase in the number of samples included in projects, which have more than doubled in the last two years alone, enabling researchers to include more conditions and better controls, as well as time courses instead of single time points. Demand has risen considerably for proteomics as software developments that help researchers to analyze and visualize data relatively easily have made it much more accessible. It is also complementary to the other omics disciplines that are often applied in clinical research such as genomics or classical biochemistry methods.
New technologies with high-throughput capabilities have created a new position for proteomics. Usually, a geonomics approach would be applied to a whole sample set, with a subset to be further analyzed with proteomics, but now, proteomics is being used to cover an entire sample set, leaving other technologies to cover a subgroup. A study at Oxford University’s Target Discovery Institute (TDI) group enabled the deep phenotyping of the host immune response in COVID-19 infections. Among the eight types of omics technologies that were used, proteomics was the best suited to stratify the COVID-19 patients into different disease severity groups. Additionally, indicators or markers were identified to help plot how the disease progresses and even allow prediction of outcomes, with the eventual aim of intervention and tailored treatment for these patients. Studies such as this can provide essential insights that will inform future drug development, clinical trial design, and personalized medicine approaches for COVID-19.
Within TDI, the Discovery Proteomics Facility research team found that using TIMS to measure ion mobility enabled high-quality 4D feature alignment in retention time, mass-to-charge ratio (m/z), ion mobility (1/K0) and intensity (Figure 1). This alignment boosted the number of quantified plasma proteins to more than 500 proteins in a single 11.5 min liquid chromatography (LC)-MS/MS (Figure 2). This depth achieved by TIMS offers new possibilities for analyzing large sample cohorts of hundreds to thousands of samples for biomarker discovery in blood plasma.
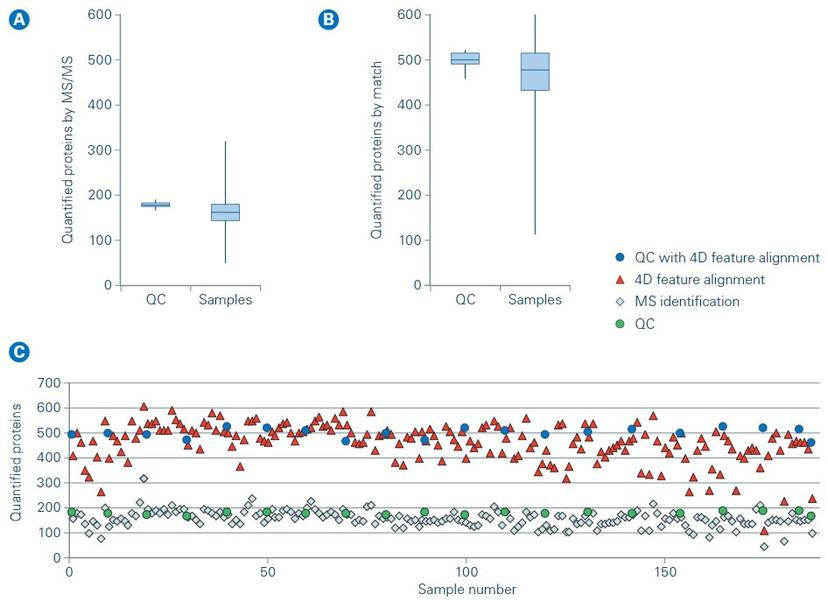
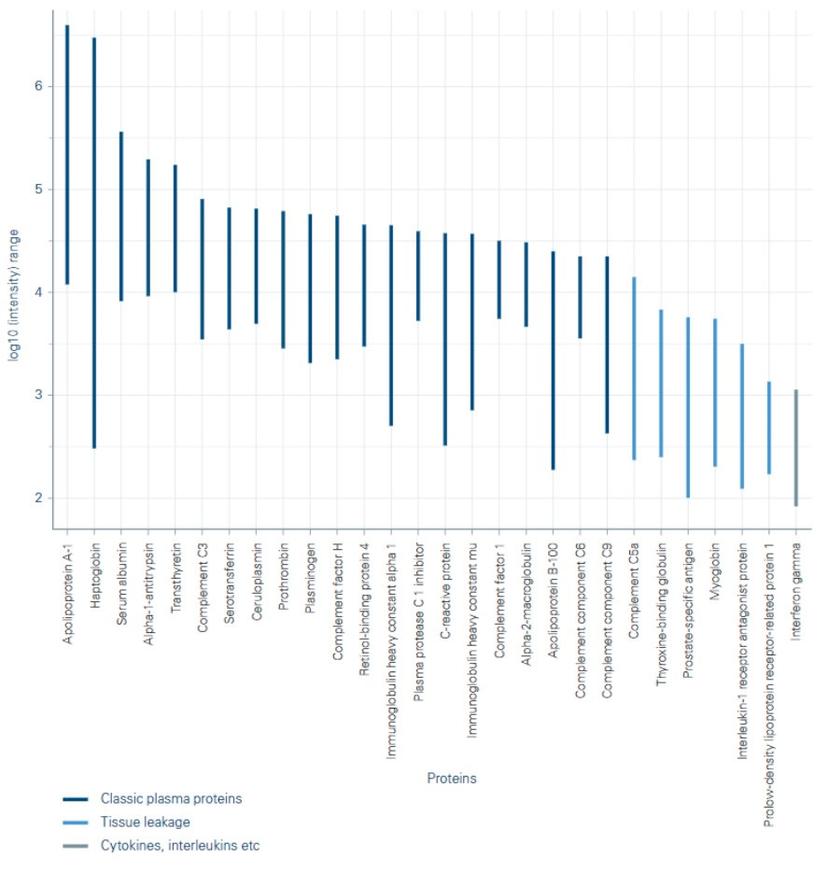
Predictions for the future of clinical proteomics
High-throughput proteomics has the potential to be used in future clinical settings. These could include, for example, installing instruments into hospitals to allow for day-to-day screening of patient samples. MS enables the measurements of multiple analytes at the same time, so proteomics could analyze a whole pathway or a whole assembly of proteins from a patient sample, potentially leading to an improved diagnosis. Another benefit is instead of being tested for one disease, many patients have comorbidities that could be identified with MS in a systems biology-based approach.
Advances in proteomics technology are set to be a major contributor to the growth in personalized medicine.
References
Grand View Research, Personalized Medicine Market Growth & Trends, https://www.grandviewresearch.com/industry-analysis/personalized-medicine-market#
Cox J and Mann M Quantitative, High-Resolution Proteomics for Data-Driven Systems Biology. Annu. Rev. Biochem. 80: 273-299 (2011).
Kosinski T, Heilig R, Bensaddek D, Bache N, Bjeld Hørning O, Fischer R, and Koch H. Plasma proteomics goes high throughput – timsTOF Pro with PASEF and 4D feature alignment to quantify 500 plasma proteins in 11.5 min. Bruker Daltonics 03, LCMS-151, 1867805 (2019).

