Cellular decision making in normal and leukemic blood stem cells
This guest editorial explores how advances in single-cell technologies to reveal novel insights into hematopoietic development
16 Aug 2024
Nicola Wilson, Senior Research Associate at the Cambridge Stem Cell Institute
Hematopoiesis, the formation of mature blood cells from blood stem cells, has traditionally been thought of as a stepwise process, and has classically been defined as a tree-like model where multipotent stem cells give rise to their progeny via an ordered process.
However, advances in single-cell technologies are challenging this classical rigid structure to reveal a more nuanced picture of heterogeneous populations, still organized in a hierarchical structure but with a more gradual progression from one stage to the next. This results in a more flexible system, which better reflects both the steady state and the adaptability required by the hematopoietic system to meet the challenges of blood demand within an organism, for example in response to external stresses such as injury or severe infection.
Understanding hematopoietic development
The Göttgens lab at the Cambridge Stem Cell Institute is using a combination of experimental and computational approaches to study how blood cells develop and how mutations within blood stem and progenitor cells can cause leukemia. Their current research focuses on early blood development, understanding blood stem and progenitor differentiation, consequences of leukemogenic mutations in leukemic stem and progenitor cells, and the computational modelling of homeostatic and perturbed hematopoiesis.
Multiomics and single-cell analysis
The Göttgens group over the last years has taken full advantage of interdisciplinary approaches and has delivered several key advancements in the application of genomics to stem cell biology. The group illustrated the power of comparative genomics in the identification of gene regulatory elements, which was followed by the first molecular characterization of enhancer elements active within hematopoietic stem cells. Construction of experimentally validated gene regulatory network models for blood stem and progenitor cells was complemented with comprehensive genome-wide transcription factor maps, describing the multifaceted nature of stem cell regulatory landscapes.
Early adoption of single-cell expression profiling techniques specifically highlighted the transcriptional heterogeneity in blood stem and progenitor cells and allowed them to deliver the first organism-scale single-cell maps for mouse gastrulation and early organogenesis. More recently, the combination of persistent labelling of stem cells with time-series single-cell genomics produced the first real-time, quantitative model of in vivo mouse bone marrow hematopoiesis.
Central to the Göttgens lab approach is the willingness to adopt new technologies and approaches to answering their scientific questions; central to this is their use of single-cell RNA-sequencing. The lab utilizes both plate-based and droplet-based single-cell RNA-seq methods, depending on the question to be answered and the cell population to be investigated. Plate-based methods are particularly useful for deep dives into specific and rare cell populations, while droplet-based approaches can capture a broader snapshot of cellular states within a tissue or population.
To enhance their understanding, the team integrates other modalities in a multiomics framework, such as ATAC-seq for chromatin accessibility and surface protein expression analysis via CITE-seq. These complementary data layers provide a holistic view of cellular state and function, enabling the lab to understand at a fundamental level the different lineage decisions and the molecular mechanisms driving these decisions.
Challenges and innovations in single-cell analysis
Despite their power, the cost of single-cell experiments can still limit the number of cells per experiment. Additionally, setting up plate-based methods is labor-intensive and can quickly become a bottleneck when working manually at scale.

To address these challenges, the Göttgens lab has chosen mosquito® HV genomics, SPT Labtech’s automated multichannel pipettor, in their RNA-seq and CRISPR screening workflows. Using true positive displacement technology, mosquito can handle liquid volumes as low as 500 nL, with speed, accuracy and precision. This has allowed the team to reduce the manual burden of single-cell workflows, increasing throughput and reproducibility, whilst maximizing the amount of data generated within the same budget.
The team uses the Illumina Nextera XT library prep kit alongside in-house reagents. While manual methods allow for miniaturization down to 4X, the mosquito system enables them to achieve 10X miniaturization, resulting in even more efficient budget use.
One of the lab's favorite features of mosquito is its user-friendly design. Unlike some traditional automation systems, mosquito’s intuitive interface allows the team to easily create and execute smaller, custom protocols as needed.
Research highlights and future outlook
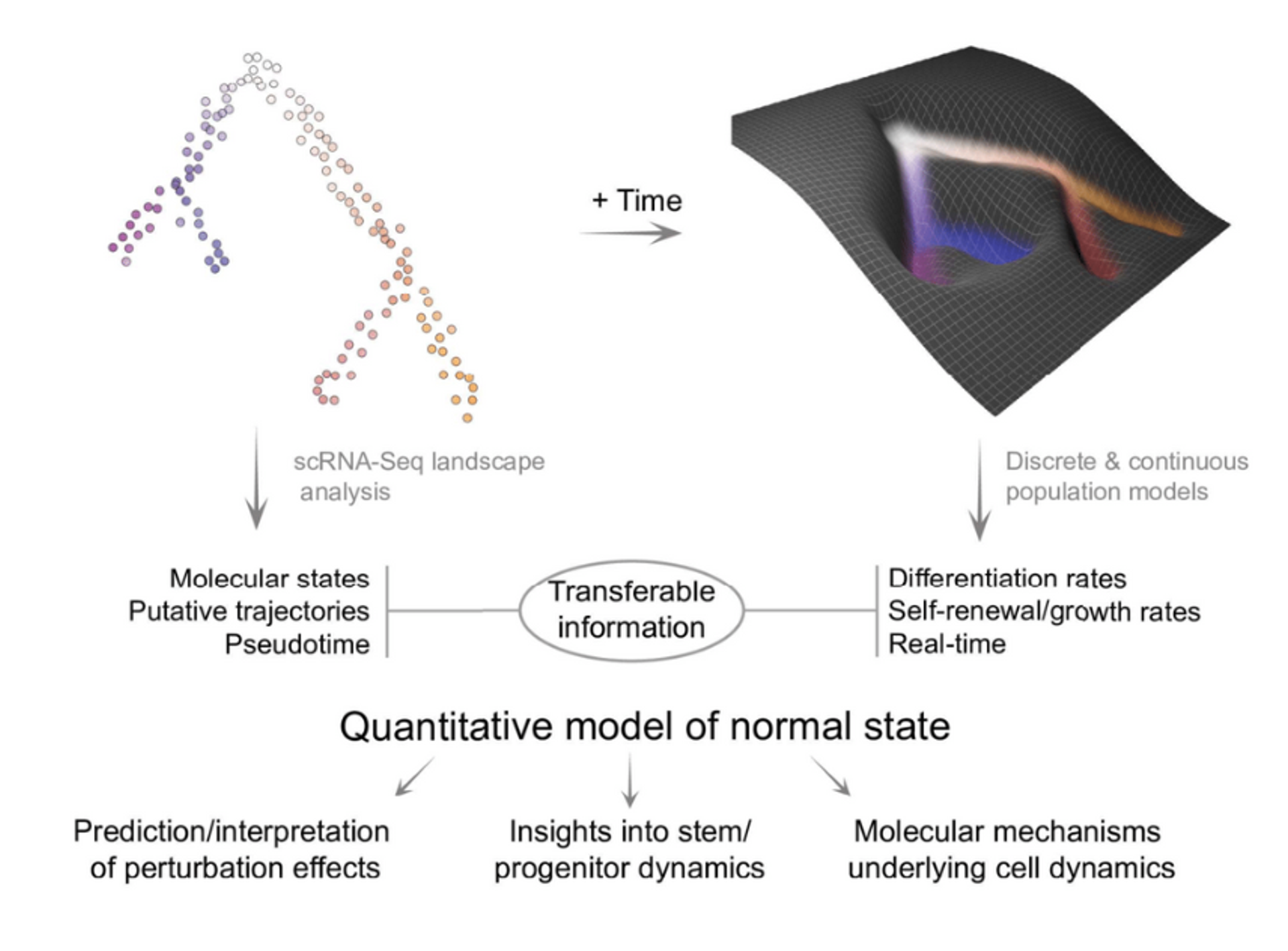
Quantitative model of hematopoietic dynamics in mouse bone marrow, describing cell behaviour with self-renewal and differentiation rates, represented as the shape of a Waddinton-like landscape. This diagram is a summary of the quantitative model and highlights the model utility and the transferable information.
A recent highlight from the Göttgens Lab is their publication in Cell Stem Cell1, where they presented, for the first time, a time- and single-cell-resolved model of murine bone marrow hematopoiesis. This study combines persistent labelling with time-series single-cell RNA-seq to create a real-time, quantitative model of tissue dynamics. This model provides new insights into the self-renewal and differentiation properties of hematopoietic stem cells and offers a framework for understanding the impact of genetic perturbations.
Looking forward, the team are working to extend their findings to human systems. This effort includes applying powerful computational models to bridge the gap between murine and human data, as well as working with in vitro systems that can maintain and amplify human hematopoietic stem cells.
This guest article was written by Nicola Wilson, Senior Research Associate at the Cambridge Stem Cell Institute.
References
1. Kucinski, I et al. (2024). A time- and single-cell-resolved model of murine bone marrow hematopoiesis, Cell Stem Cell, 31 (2), 244-259. https://doi.org/10.1016/j.stem.2023.12.001

