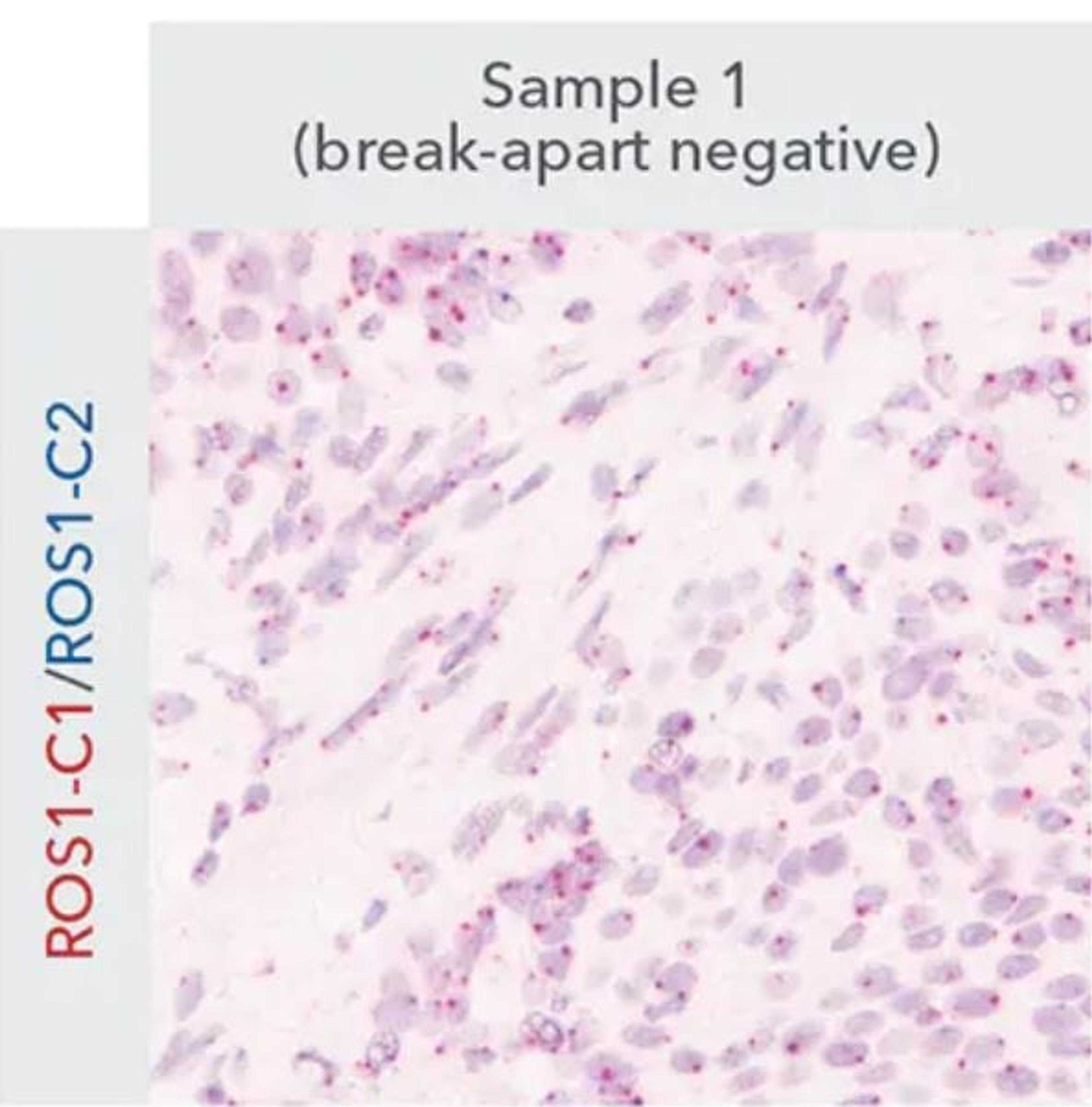
High-resolution detection of DNA copy number and structural variants with novel ISH technology
Get an introduction to ACD’s novel chromogenic in situ hybridization (ISH) DNAscope Assays
21 Feb 2022

Introducing ACD's novel chromogenic in situ hybridization (ISH) DNAscope Assays for high-resolution detection of DNA copy number and structural variations. DNA ISH is a spatial profiling tool based on hybridizing labeled complementary probes to the target DNA inside the cell nucleus to visualize any genetic alterations in tissues.
In this on-demand SelectScience® webinar, learn how current limitations of the DNA ISH assay, such as limited morphological detail, difficult interpretation and low-resolution detection due to larger probe sizes are addressed by the new assay. The DNAscope chromogenic duplex (red/blue) staining allows researchers to use a standard brightfield microscope to visualize and quantify gene copy number variations (amplifications/deletions) and gene rearrangements or fusions in tissues with spatial and morphological context at single-cell resolution.
Watch on demandRead on for highlights from the live Q&A article or register to watch the webinar at a time that suits you.
Do you offer any control probes for DNAscope assays?
VM: Control probes to assess tissue quality are not needed. However, our copy number variation application does have the chromosome probes which can serve as an internal control.
Is it okay to use a longer or shorter probe incubation time than 16 hours?
VM: Yes. The overnight probe incubation is meant to serve as a convenient breakpoint in the assay, so the assay can be split over two days, and it's okay to go a couple of hours below or above the 16 hours. We recommend not exceeding 18 to 20 hours for risk of seeing background noise.
What is the cut-off for determining a sample as positive?
VM: The inauguration guidelines that we provide are meant to serve as recommendations for interpreting your DNAscope signal. The criteria for scoring or setting a cut-off should be really determined by the end user through appropriate validation studies in their own sample cohorts.
Is the double-Z basic mechanism still in place to ensure specificity?
VM: Yes. The DNAscope probe design is still based on our proprietary double-Z design to ensure specificity.
Would it work in the case of plants, for example, in CRISPR experiments?
VM: We have optimized the DNAscope assay for FFP tissues. Plant tissues would require optimization from the user's end in terms of the pre-treatment to get optimal detection. And secondly, we would also need to determine probe design feasibility by our informatics team.
Can we design custom probes for any target, or are we limited to the target shown in this presentation?
MN: Yes, we can custom design any target probe. We would need to perform feasibility for those targets, probe design feasibility. Our recommendation would be that you would send that across to us and then we can get back to you.
But, no, they are not limited to the ones that we showcased in this presentation. Those are merely to show the performance of the assay, and we can design a custom probe for any target of interest.
Can DNAscope assays be combined with your RNAscope or BaseScope assays?
MN: At this point, we have not done any feasibility on combining the DNAscope assays with the other assays. That is something that we can keep in mind as we continue to improve our products in order to cater to the scientific questions that are being asked. At this point, we have not validated the combination of these assays.
Do you also have control slides for each of the probes in the catalog?
MN: We do not have control slides for the probes, we do have control probes, particularly for copy number variations. These are the CEP control probes that would enable us to figure out the standard enumeration of the target, and the chromosomes that we expect the gene to be in. Other than that, we do not have any control slides.
As an assay control, we typically have found that the researchers would have different orthogonal methodologies by which they would have validated either the break apart and/or the copy number. And we are presenting a spatial analysis tool to get to those key applications at this point.