How IDT’s new HiFi Cas9 mutant is helping to overcome challenges in the clinical application of CRISPR technology
Dr. Chris Vakulskas talks about the work he is doing with CRISPR, the challenges involved and new innovations within the field
19 Nov 2019
Dr. Chris Vakulskas is Director of Enzyme Evolution at Integrated DNA Technologies and leads the protein engineering and novel technologies innovation group at IDT that works heavily with the latest CRISPR technologies. In this SelectScience interview, Vakulskas explores CRISPR’s capacity for high-specificity gene editing, its economic viability and the impact CRISPR will have on personalized medicine and gene therapy.

Vakulskas highlights some of the challenges involved in transferring this research to clinical applications, such as the delivery of the system to its intended target, the potential consequences of off-target editing and the effects CRISPR may have on adaptive immunity. He also outlines IDT’s newly developed HiFi Cas9 mutant and explains how it can overcome some of the challenges discussed, bringing routine therapeutic genome editing closer to reality.
Please introduce yourself and your role at IDT
My name is Chris Vakulskas and I am the Director of Enzyme Evolution at Integrated DNA Technologies (IDT). I presently lead the protein engineering and novel technologies innovation group at IDT. We use basic bacterial genetics and directed evolution to isolate novel protein mutants that have either enhanced or novel functionality. We currently focus heavily on new CRISPR technologies, with an emphasis on Cas9 from S. pyogenes and Cas12a (Cpf1) from Acidaminococcus species. I lead an exceedingly talented group of scientists that can do everything from saturated mutagenesis screening in E. coli to transient transfection of CRISPR ribonucleoprotein complexes into immortalized human cells. Many of our efforts and throughput are enhanced by the use of liquid handling robots, next-generation sequencing, and above all, compartmentalization of genotype-phenotype in individual bacterial cells.
How did you become interested in CRISPR research?
Most of our most fundamental molecular biology tools come from bacteria and the viruses that infect them (bacteriophages). CRISPR is part of a primitive bacterial immune system that is used to protect from phage and other foreign DNA that would disrupt genomic integrity. My interest has always been in bacterial physiology and protein biochemistry, and CRISPR is just one more example.
What important implications do CRISPR systems have for personalized medicine?
In my view, the main benefit of CRISPR is that it can be both highly-specific and (unlike Transcription Activator-Like Effector Nucleases (TALENs) or Zinc Finger Nucleases (ZFNs)) economically targetable. One person may have a hereditary condition caused by a unique mutation, and CRISPR eliminates much of the expense that is associated with the targeting mechanism. With previous technology, expensive and laborious protein engineering had to take place in order to provide specific targeting of the DNA cleavage machinery. With CRISPR, all you need to do is synthesize a unique RNA and combine it with a universal protein component (Cas9, Cas12a, etc.).
How close are we to translating this to the clinic? What are the main challenges of existing systems?
The main challenge for any drug or therapeutic remains one of delivery. How do you get the treatment to the target tissues in a safe manner and ensure that those reagents stay in the tissue long enough to affect the desired outcome? The low hanging fruit of CRISPR are so-called ex vivo therapies, wherein a patient’s own cells can be removed from the body, treated with CRISPR, and transplanted back into the person. These treatments are very close to becoming a reality (thanks to Integrated DNA Technologies’ (IDT’s) HiFi Cas9!) but effective delivery into specific tissues within the human body remains a significant barrier.
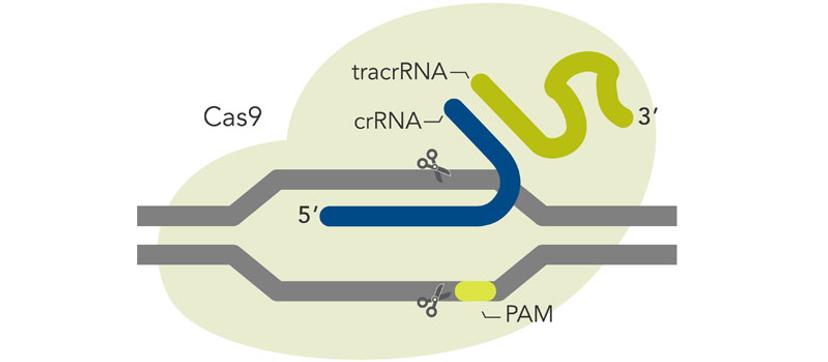
Off-target effects – can we really predict this in non-human pre-clinical models?
We cannot routinely predict the off-target sites for a given Cas9 target sequence….yet, although these predictive algorithms are seemingly getting better. What we can do, however, is to use delivery methods like RNP coupled with HiFi Cas9 to minimize the chance for off-target editing and maximize safety. In a therapeutic context, any remaining off-target editing can be evaluated using known procedures like GuideSEQ coupled to multiplexed amplicon sequencing technologies like rhAmpSeq.
In vitro culture – serum is known to affect cellular behavior over time, does this have an effect on CRISPR studies? How might more complex 3D cellular models improve research?
I think most people are viewing many of the ex vivo therapies as one or two treatment cures, so this is probably not a concern or relevant.
What about human adaptive immunity to Cas9?
While it has been shown that humans have existing immunity to Cas9 from both S. pyogenes and S. aureus, this is probably more a safety consideration rather than a barrier. Most ex vivo treatments in clinical trials do not rely on multiple treatment regimens, therefore the opportunity for a meaningful harmful immune response is negligible.
How does IDT overcome these solutions with its new HiFi technology? What is the new HiFi Cas9 enzyme you’ve developed, how does it work, and why is it better?
The HiFi Cas9 enzyme is a mutant of the wild-type (WT) Cas9 protein that dramatically reduces off-target editing without compromising on-target potency. Most existing high fidelity Cas9 mutants reduce off-target editing at the expense of overall on-target potency. We selected for mutations that had little effect on on-target potency using directed evolution in bacteria with a specific emphasis on maintaining on-target potency.
Since its discovery, and publication of the Nature paper in 2018, what type of work is currently being done with this HiFi Cas9 enzyme mutant, or others?
Internally, we are always looking to improve our products, which includes discovering the next “version” and also looking for different combinations of these mutations and contexts that allow people to more effectively use these enzymes in “cell-free” applications where nuclear targeting is unimportant, and the trade-off between on/off-target potency is not the same as it is in living cells. Externally, HiFi Cas9 is becoming a standard for therapeutic genome editing as it is available in an “off-the-shelf” cGMP form from our commercial partner Aldevron and has demonstrated utility in multiple high-tier publications and clinical trials.
How long do you predict it will be until CRISPR is being used routinely as a form of genetic therapy?
I think for certain diseases we’re looking at a 5-10 year timeline at most. For more complex diseases, we still have not solved the major problem of delivery that has always existed and it’s really an interesting and exciting open question.
