Microbiome research can benefit from breath biomarker analysis
Harness exhaled breath for real-time monitoring of microbial metabolites and health impacts
8 Aug 2023Biomedical research has been transformed by the discovery that the human body is colonized by microorganisms (termed the microbiota, or the microbiome), which can dynamically interact with the many tissues, organs, and systems of the body to impact health and the development of disease.
The community of microbes within the body have unique metabolic processes, and the metabolites they produce are thought to be the key to their interaction with us, and the mechanism through which they impart their effect on the body. Typically, the study of the microbiome involves metagenomic analysis or the analysis of microbially-produced metabolites in waste products such as fecal matter and urine. Exhaled breath is underutilized in microbiome research, despite the power it holds to offer different, yet complementary insight. Like urine and fecal matter, breath is a waste product from the body that is enriched with informative compounds that can provide insight into the function of systems within the body (Figure 1). However, unlike traditionally utilized biological samples, a large volume of breath can be collected in rapid succession - it has been estimated that we take an average of 22,000 breaths per day1, with the volume of a single exhaled breath at rest being approximately 400 – 500 mL2.
What compounds are in inhaled breath?
Breath is comprised of a diverse matrix of compounds originating from both outside and inside of the body, with compounds generated from within the body capable of diffusing away from their point of origin into the bloodstream. Once in the circulatory system, these compounds travel through to the lungs, whereby they can pass from the alveolar surface into the air to be exhaled. In order for a compound to be able to do this, it must be volatile, meaning it is gaseous at room temperature. Research so far has primarily focused on volatile compounds produced by human metabolic processes, and immune-related processes such as inflammation. However, there are also volatile compounds in the breath that originate from the microbiota, which could provide a new platform through which to study their interaction with us. Critically, many of the already well-studied microbially produced metabolites are volatile, and therefore can be identified and quantified in exhaled breath across a variety of experimental conditions.
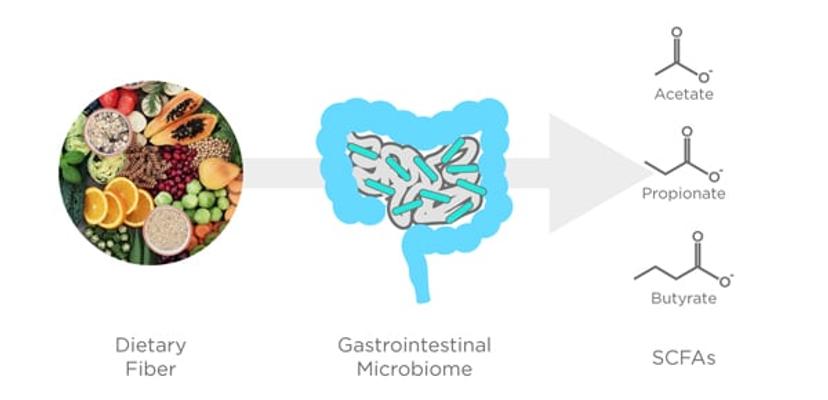
There is a diverse assortment of volatile compounds detectable in breath from a range of chemical classes including alcohols, hydrocarbons, fatty acids, and many more. Some of the most abundant volatile metabolite groups produced by the microbial community in the colon are the short-chain fatty acids (SCFAs), which are produced from the fermentation of dietary fiber (Figure 2). The SCFAs have many complex roles in the regulation of the healthy gut environment by modulating barrier function, the immune system, providing an energy source, and more3,4. Three SCFAs in particular, acetate, propionate, and butyrate, make up 90-95% of the SCFAs produced by the microbiota in the colon, and there many exciting links have been made between the relative abundance of SCFAs in the body and the beneficial metabolic effect they have, as well as their correlation to reducing the incidence of certain diseases, including colon cancer5,6. Crucially, these three well-studied SCFAs are volatile, and are readily able to be measured in exhaled breath, opening the possibility of analyzing the effect of the SCFAs and other volatile microbial biomarkers on health and disease through the analysis of breath7.
Breath tests can provide real-time insights into the activity of the microbiome
Hydrogen and methane breath tests (HMBTs) that probe the microbiome in the gastrointestinal tract are already readily available and demonstrate the potential of breath biomarkers by enabling the diagnosis of conditions such as small intestinal bowel overgrowth (SIBO), and carbohydrate malabsorption (CM) conditions such as lactose intolerance. During an HMBT, the patient is asked to fast for a period of time (usually 12 hours), and then ingests a test substrate depending on the particular test, For SIBO testing for example, a solution of lactulose diluted in water is ingested. Next, the breath is either analyzed with an at-home breath testing device, or collected over a few hours, and the samples analyzed in a laboratory to assess the changes of hydrogen and methane over time (Figure 3). Therefore, breath tests that measure a wider range of volatile compounds could serve as biomarkers for various processes relating to the microbiome, enriching clinical research with quantifiable markers closer to real-time information about the changes in microbial metabolism in the body.

The HMBTs operate on the basic principle of ingesting a test substrate and measuring the response of relative compounds in the breath to make biological inferences about the microbiome. This approach is already being successfully developed to probe liver function. The exogenous (EVOC) probe limonene can be used to probe liver function through its metabolism by the CYP450 enzymes, and so offers a promising non-invasive alternative to liver biopsies for diagnosing cirrhosis and earlier-stage liver disease8,9. A similar approach with other compounds to probe microbial enzymatic pathways can be developed, opening up the possibility for the next-generation complex monitoring of patients’ microbial communities.
Breath biomarker analysis is a complementary platform to fecal analysis
The current method of choice for many microbiome studies is fecal analysis. Fecal matter contains informative compounds and live microbiota that can be identified and quantified. However, there are several methodological limitations to fecal sampling that breath can circumvent, and therefore complement the information gained. Fecal matter reflects a broad period of digestive passage, the average whole gut transit time in a healthy person ranges from 10 to 73 hours10, meaning that all information gained lags at least several hours behind the dynamic processes of the gut. Through HMBTs, the lag-time between the production of gas by the microbiota in the gut, and the detection of the gas in the breath can be quantified – as one study directly infused lactulose into the caecum, and the time for fermentation by bacteria, and subsequent detectable hydrogen in the breath was five minutes or less11.
Mucosally adherent microbes, especially those in the small intestine are commonly missed, and environmental changes as a result of passage may lead to misrepresentative microbial composition12. This makes location-specific microbiome changes difficult to assess via fecal sampling, for example in conditions such as small intestinal bacterial overgrowth. Some informative compounds such as SCFA butyrate are also metabolized by colonocytes, therefore, the concentrations that end up in fecal matter may be unrepresentative of the physiological levels in the gastrointestinal tract. Breath analysis on the other hand allows for the monitoring of metabolites with only a short lag-time after they are produced, and which are not biased for specific locations in the gut. Sample variability can be easily introduced during the handling of liquid samples of urine or feces, as highly volatile compounds in the air away from detection equipment can be lost.
Breath analysis can be incorporated in microbiome research in many innovative ways
On top of providing a complementary analysis that overcomes the specific limitations of fecal sampling, there are several unique bonuses of breath that make it promising as another tool in the arsenal of microbiome research. A large volume of breath can be taken regularly, allowing for longitudinal monitoring of a wide range of metabolic and biochemical processes of interest. This benefit is best utilized in clinical trials, where biomarkers for metabolic pathways could be monitored in real-time in response to treatments. Breath biomarkers can allow for faster endpoint decisions and minimize the costs of lengthy clinical studies, as well as reduce the need for expensive invasive monitoring. Another benefit of breath sampling to clinical trials is that it is inherently non-invasive, with breath leaving the body as a waste product almost continuously. This can improve patient or participant comfort to make ethical approval easier, and uptake during recruitment is likely to be higher.
The possibilities for innovative study designs that utilize breath analysis for a deeper understanding of the microbiome are endless. Microbially-produced volatile compounds could be analyzed between populations with different microbial communities to investigate how the metabolites change, across disease cohorts to measure how metabolite concentrations could be potential diagnostic biomarkers. Certain breath compounds could also serve as efficacy markers of an intervention such as assessing the impact of food ingredients such as fibers, pro- and pre-biotics, and fermented foods on digestive health, as well as the response to different therapeutics. Trials that incorporate both fecal and breath sampling are powerfully for multi-dimensional analysis that could provide the next-generation insight to revolutionize how we understand the microbiome, and the impact the microbiome has on our health.
This article was originally shared and written by Owlstone Medical. Learn more about its Owlstone Breath Biopsy® Platform >>
References
1. Breathing | Canadian Lung Association [Internet]. [cited 2023 May 25]. Available from: https://www.lung.ca/lung-health/lung-info/breathing
2. Pleil JD, Wallace MAG, Davis MD, Matty CM. The physics of human breathing: flow, timing, volume, and pressure parameters for normal, on-demand, and ventilator respiration. J Breath Res. 2021 Sep 27;15(4):10.1088/1752-7163/ac2589.
3. Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de los Reyes-Gavilán CG, Salazar N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front Microbiol. 2016 Feb 17;7:185.
4. Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002 Dec 17;217(2):133–9.
5. Cronin P, Joyce SA, O’Toole PW, O’Connor EM. Dietary Fibre Modulates the Gut Microbiota. Nutrients. 2021 May 13;13(5):1655.
6. O’Keefe SJD, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015 Apr 28;6:6342.
7. Lee JH, Zhu J. Analyses of short-chain fatty acids and exhaled breath volatiles in dietary intervention trials for metabolic diseases. Exp Biol Med (Maywood). 2021 Apr;246(7):778–89.
8. Ferrandino G, Orf I, Smith R, Calcagno M, Thind AK, Debiram-Beecham I, et al. Breath Biopsy Assessment of Liver Disease Using an Exogenous Volatile Organic Compound—Toward Improved Detection of Liver Impairment. Clinical and Translational Gastroenterology. 2020 Sep;11(9):e00239.
9. Ferrandino G, De Palo G, Murgia A, Birch O, Tawfike A, Smith R, et al. Breath Biopsy® to Identify Exhaled Volatile Organic Compounds Biomarkers for Liver Cirrhosis Detection. Journal of Clinical and Translational Hepatology. 2023 Jun 28;11(3):638–48.
10. Lee YY, Erdogan A, Rao SSC. How to assess regional and whole gut transit time with wireless motility capsule. J Neurogastroenterol Motil. 2014 Apr 30;20(2):265–70.
11. Read NW, Al-Janabi MN, Bates TE, Holgate AM, Cann PA, Kinsman RI, et al. Interpretation of the breath hydrogen profile obtained after ingesting a solid meal containing unabsorbable carbohydrate. Gut. 1985 Aug;26(8):834–42.
12. Allaband C, McDonald D, Vázquez-Baeza Y, Minich JJ, Tripathi A, Brenner DA, et al. Microbiome 101: Studying, Analyzing, and Interpreting Gut Microbiome Data for Clinicians. Clin Gastroenterol Hepatol. 2019 Jan;17(2):218–30.

