Nanolive launches EVE Analytics for the CX-A: A complete solution for live-cell imaging and analysis
Discover the automated, quantitative, and label-free software-hardware integration set to accelerate therapeutics research
17 Mar 2021
Nanolive SA has announced the market launch of an integrated and user-friendly software solution for unbiased, non-invasive, continuous live-cell analysis and quantification: EVE Analytics.
The cell is the basis of all life on earth, thus the capacity to interact with living cells holds the key to making great discoveries and advances in medical science.
Nanolive’s CX-A together with its quantification software EVE Analytics, represents a significant breakthrough in the field of quantitative live-cell imaging and analysis.
The company's mission is to use its proprietary, non-invasive, unbiased, live-cell data to revolutionize the understanding of living cells and accelerate the discovery and development of therapeutics. With Nanolive’s platform, researchers get higher significance, earlier and faster.
Live-cell imaging for the pharmaceutical industry
Live-cell imaging is a powerful tool in drug discovery because it enables researchers to quantify and compare the dynamic responses of unperturbed cells with those of drug-perturbed cells, providing novel mechanistic insights into the responses of cells to a candidate drug. With Nanolive’s technology, the data output is richer and more reliable because information can be collected from multiple sub-cellular compartments simultaneously (multiplexing) in a totally non-invasive fashion, meaning results are not compromised by phototoxicity and/or photobleaching. Nanolive’s label-free technology makes it possible to image cells for long periods of time, at high spatio-temporal resolution, without signal deterioration. Furthermore, Nanolive’s imaging technology grants full quantitative analysis. The quantity and complexity of the images generated allow us to visualize biological processes in unprecedented detail, but also magnifies the challenges associated with image analysis.
State-of-the-art of cell segmentation
Cell segmentation relies on one simple but fundamental concept; cells must have a signal that is different from the background. To achieve this, most studies use contrast agents, which bind to specific molecular components of the cell, allowing cells to be differentiated from the background by simple thresholding. The most common contrast agent is the nuclear stains DAPI, but this dye requires the sample to be fixed. Other nuclear dyes can be tolerated by living cells, but not for long, as their presence perturbs fundamental genetic processes, and they require excitation to generate fluorescence, which generates substantial phototoxic stress. Nuclear dyes make it possible to quantify the number of cells but do not provide a lot of information about the size/area/shape of the cell. Using cytosolic stains overcomes this limitation but complicates analysis for two reasons. Firstly, the local quantity of cell material they stain varies, and so the fluorescent signal intensity differs dramatically within a cell. Secondly, the variability in cell edges morphologies makes it much more complex to separate touching cells, prohibiting a simple thresholding approach like the one applied to segment nuclear objects.
EVE Analytics
EVE Analytics from Nanolive benefits from a new segmentation technique, developed specifically for Nanolive’s label-free images.
Nanolive images are different from other traditional label-free images: they are richer and more complex. The complexity of the signal distribution and the texture of the image disturb the traditional detection of the center of the cell or of any other signal that could serve as a seed. It is especially difficult when cell confluence increases, and the cells begin to touch. Nanolive’s application development team led by Dr. Mathieu Frechin, head of AI for quantitative biology, managed to circumvent this problem and this is where the true innovation of its newest product EVE Analytics started.
“Thanks to EVE Analytics we can now offer precise cell segmentation of living cells for virtually unlimited periods of time, which is a significant breakthrough in the field of live cell imaging” stated Dr. Frechin about this achievement.
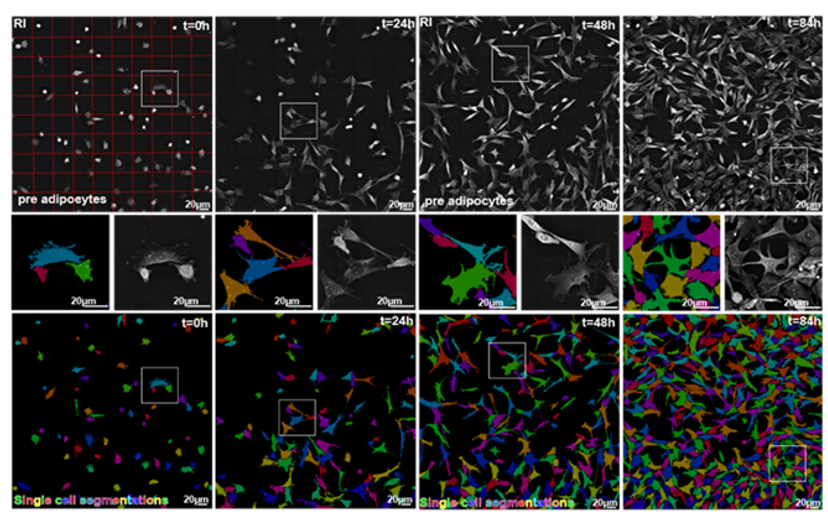
Primary object detection relies on signal preparation and advanced processing methods to detect the cellular object seed. This is then followed by a secondary step based on a refractive index (RI) signal, which allows capturing the fine boundaries of cellular objects. EVE Analytics works over long-term experiments where cell confluency changes significantly with no reduction in quality. In the figure below, the zoomed-in images show the high-precision of the segmentation. The fine details of the cell membrane protrusions (filopodia and lamellopodia) are captured, while touching cells are accurately delineated, proving that there is no tradeoff between scale and precision.
Moreover, EVE Analytics simultaneously calculates a multitude of live cell metrics, such as cell count; confluency; cell ellipticity; cell circularity; cell perimeter; cell area; average dry mass; RI; cell compactness; dry mass density; cell extent; total dry mass.
On top of this, thanks to the hard work of Nanolive’s R&D and software team the EVE Analytics software is extremely easy to use and does not require any prior knowledge nor special training.
Want the latest science news straight to your inbox? Become a SelectScience member for free today>>
