Navigating the complexities of spatial biology
Join experts as they discuss the significant hurdles in spatial proteomics and how innovative tools such as the EVOS S1000 Spatial Imaging System are set to redefine research capabilities in this rapidly evolving field
7 Aug 2025

Charlotte Stadler, Co-Director of the Spatial Proteomics Infrastructure Unit at KTH/SciLife Lab in Stockholm, Sweden
Spatial biology and spatial proteomics are revolutionizing our understanding of complex biological systems by highlighting the intricate spatial relationships between proteins, cells, and tissues. This innovative approach allows researchers to visualize and analyze the distribution and interactions of biomolecules, leading to a deeper understanding of cellular functions, disease mechanisms, and therapeutic responses.

Juan J. García-Vallejo, Associate Professor at the Department of Molecular Cell Biology and Immunology at Amsterdam UMC
Despite these exciting advancements, several challenges hinder the full potential of spatial proteomics. Researchers face obstacles such as the need for standardized methodologies, data management issues stemming from the vast amounts of information generated, and the demand for advanced imaging technologies capable of capturing the nuances of biological samples. As these challenges mount, the need for cutting-edge instruments such as the Invitrogen™ EVOS™ S1000 Spatial Imaging System* from Thermo Fisher Scientific becomes increasingly critical to facilitate research and enhance outcomes in translational and clinical applications.
We spoke with Charlotte Stadler, Associate Professor at Department of Protein Sciences at KTH and Head of the Spatial Proteomics Infrastructure Unit at Science for Life Laboratories in Stockholm, Sweden, and Juan J. García-Vallejo, Associate Professor at the Department of Molecular Cell Biology and Immunology and Director of the Microscopy and Cytometry Core Facility at Amsterdam UMC, to discuss the current challenges in spatial biology, particularly in spatial proteomics. We also inquired about the innovative tools required to address these hurdles and drive further advancements in the field.
Leading the way in spatial biology
Charlotte Stadler leads a team of researchers, research engineers and Ph.D. students, providing essential services to the life sciences community. “Throughout my research career and now also my infrastructure career, my focus is on spatial biology and spatial proteomics, where we primarily use antibodies to explore protein expressions in cells and intact tissue sections through multiplexed imaging, ” she explains. Her own research focuses on clinical and translational studies conducted in collaboration with clinicians, leveraging spatial omics methods to improve patient care in real-world settings.
In addition to her research, Stadler’s team provides services to the broader life sciences community, “We also provide services to the life science community using advanced spatial omics methods to allow more researchers to do work that otherwise might not be possible.” This collaborative approach drives innovative projects, enhancing the understanding of protein functions in both healthy and diseased tissues, particularly in cancer research. She explains, “Everyone is realizing the advantages of looking at different types of omics and intact tissue sections, but I would say that it's still very expensive. For that reason, it is not yet applied to every lab. I think this is one of the reasons why we’ve invested a lot of funding to make these methods available within the research infrastructure so that we can help others to still get access to the technologies.”
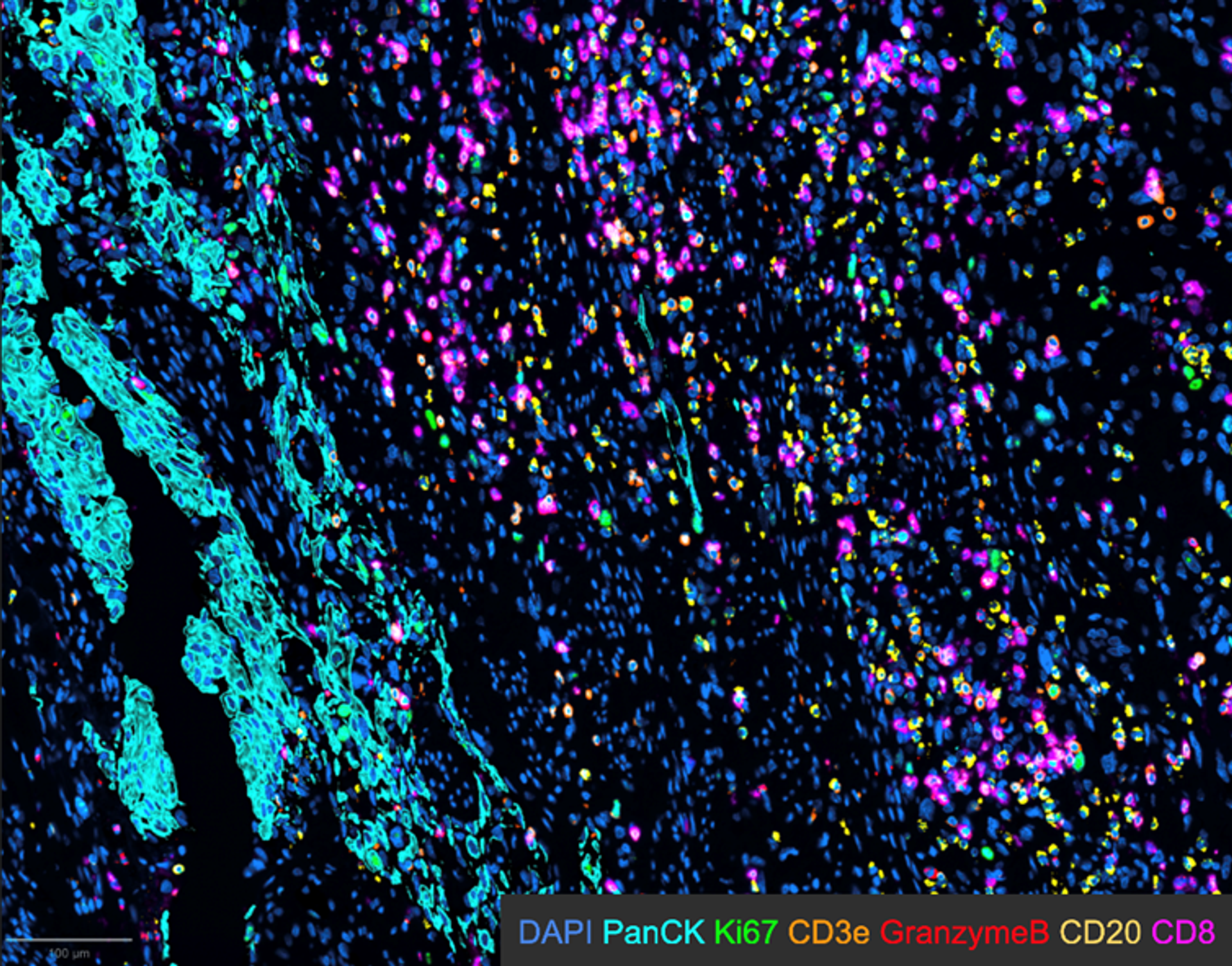
The image is from a bladder cancer tissue section, with defined tumor areas from epithelial origin marked by PanCK (pancytokeratin - cyan), various immune cells (CD3, CD8, CD20) and some functional/proliferation markers (granzyme B, Ki67). Image acquired by Maria Lung. The image was obtained using the EVOS S1000.
García-Vallejo shares insights from his dual role in academia. “I am a medical doctor by training, and I do research in the field of immunology. My research group focuses on immune monitoring of complex diseases to identify biomarkers that will help us predict disease outcomes and better understand the physiopathology of multiple diseases.”
García-Vallejo also serves as the Director of the Microscopy and Cytometry Core Facility, which he founded in 2016. “Right now, it provides a service with a team of about 10–12 operators to a community of more than 500 researchers here at Amsterdam UMC, ”he explains. The facility’s services primarily focus on immunophenotyping based on multiplexed methodologies. He continues, “Our services cover or focus especially on multiplexed imaging both at the single-cell suspension level, so for that we use all kinds of flow cytometry methods, but also on tissue—that is really our strength. ”
García-Vallejo highlighted the rapid increase in demand for multiplexed imaging at the Amsterdam UMC's core facility. To meet this demand, the facility has quickly expanded both its equipment and data analysis capabilities. Additionally, García-Vallejo has been beta testing the EVOS S1000 spectral imaging technology from Thermo Fisher Scientific , which he is particularly excited about, as he believes it will significantly accelerate research in multiplexed tissue imaging.
Navigating the challenges
Spatial biology is rapidly evolving, driven by advancements in imaging technologies and molecular profiling. This evolution enables the simultaneous analysis of multiple protein markers within tissue sections, marking a shift from traditional techniques. Spatial omics combines these methods with spatial resolution, allowing researchers to examine biomolecule distribution and interactions in their native contexts, leading to deeper insights into cellular functions and disease mechanisms.
I think there are mainly three problems that are really hampering development. The first one would be the availability of reagents that have been validated properly. All these technologies are based on antibodies and require significant optimization, particularly when you work with tissues.
Juan J. García-Vallejo, Associate Professor Department of Molecular Cell Biology and Immunology at Amsterdam UMC
García-Vallejo shares his perspective on the bottlenecks in the field. “I think there are mainly three problems that are really hampering development. The first one would be the availability of reagents that have been validated properly. All these technologies are based on antibodies and require significant optimization, particularly when you work with tissues.” He emphasizes that without the right clones or protocols, even the best antibodies will not yield good results.
He also points out challenges associated with multiplexed imaging: selecting the right clones and antibodies and managing signal-to-noise issues. While good resources exist for antibody selection, “the time it takes to really test and validate and optimize, delays a lot the process of coming up with a multiplexed panel solution. ” Some technologies, while enhancing the signal, introduce lengthy staining procedures. García-Vallejo suggests that in an ideal scenario, solutions with directly labelled antibodies could drastically reduce staining time, from a week to just a few hours, accelerating the generation of reliable data.
García-Vallejo identifies data analysis as another major bottleneck in multiplexed imaging. He explains that there is, “not one solution that fits all problems, ” forcing researchers to navigate multiple platforms and transfer data back and forth. This process involves correcting signal overlap, eliminating noise, and performing segmentation to identify single cells within tissue.
Stadler also shares challenges she has encountered. “I think that since this is such a rapidly evolving field, standardized workflows are still not really there. Every project requires some hands-on work and custom solutions.” Additionally, she notes the challenges posed by the large amount of data being generated, leading to logistical and practical problems with data management. “That's the challenge, but also an opportunity, I think, the data-driven aspect of it, ” she says.
Always seek expert advice
For those new to spatial biology and imaging, García-Vallejo emphasizes the importance of leveraging available resources. He advises, “Look for your nearest core facility that specializes in the topic, and reach out to experts who have been doing this work. Establishing collaborations can significantly lower the threshold for entering the field.”
García-Vallejo also recommends starting with smaller projects to avoid feeling overwhelmed by complexity. “Don’t jump straight into a 35-color panel, begin with simpler tasks and gradually build your expertise,” he suggests. This incremental approach will help newcomers gain confidence and skills as they explore the rapidly evolving landscape of spatial biology.
Stadler emphasizes the importance of clearly defining research questions when embarking on spatial biology projects. She states, “when generating large image datasets with numerous parameters, it's crucial to think about your main hypothesis or question. This will guide the project design and influence how many samples you need. Unless you are after a data driven project approach where unsupervised analysis of many markers are used to explore the samples.”
How the EVOS S1000 supports advanced spatial biology
The EVOS S1000 Spatial Imaging System is an advanced instrument that tackles key challenges in spatial biology and multiplexed imaging. Designed to streamline the tissue imaging process, the EVOS S1000 allows researchers to simultaneously detect biomarkers using a multiplex approach with fluorescence and brightfield imaging in a fast, yet accessible manner.
The EVOS S1000 instrument was a very good fit in our lab because it complements the instrumentation that we already have. It allows us to much faster image our preliminary results and data that we generate, speeding up our workflows.
Charlotte Stadler, Co-Director Spatial Proteomics Infrastructure Unit at KTH/SciLife Lab in Stockholm, Sweden
Stadler explains, “For us, the EVOS S1000 instrument was a very good fit in our lab because it complements the instrumentation that we already have. It allows us to much faster image our preliminary results and data that we generate, speeding up our workflows.”
She also highlights its flexibility and efficiency, “Thanks to the EVOS1000, we can be more spontaneous and progress faster. The instrument is very fast compared to some imaging set-ups that we have used before.” The system also streamlines downstream processing, reducing the steps needed after imaging. “It's also a relatively cheap solution for us compared to some of the other instruments used for longer imaging runs,” she adds.
García-Vallejo also recognizes the potential of the EVOS S1000 in multiplexed imaging. He believes that the system will help researchers accelerate data generation, streamline workflows, and improve overall research efficiency, allowing them to focus on scientific questions rather than technical limitations.
Looking ahead
As spatial biology evolves, tools such as the EVOS S1000 Spatial Imaging System will play a critical role in driving the next wave of discoveries and innovations. The integration of advanced imaging systems promises to address current challenges, enabling researchers to gain deeper insights into cellular interactions and therapeutic responses. By fostering collaboration and concentrating on specific research goals, scientists can leverage these advancements to significantly enhance personalized medicine and beyond.
*For Research Use Only. Not for use in diagnostic procedures.

