Organ-on-chip technology transforms infectious disease modelling
Innovative 3D in vitro microphysiological test systems improve the understanding of host-pathogen interactions and the evaluation of antimicrobial treatments, a potential aid in the fight against infections and resistance
21 Jun 2024
The 21st century has been characterized by various infectious diseases, most recently the COVID-19 pandemic, which has had a devastating impact on people and their livelihoods around the world. With a sense of urgency, the scientific community tried to understand the infection in detail and to find intervention options as quickly as possible.
Representative models of the infection process have been a major bottleneck in this process for biomedical research and lead optimization in preclinical drug development. Recently, innovative 3D in vitro microphysiological test systems known as organs-on-chip (OoC) have emerged as a promising approach to bridge this gap. In 2021, such models demonstrated their potential for studying the infection and inflammation process in SARS-CoV-2 disease¹. However, this is only one of many studies of infection processes using OoC, and this article aims to introduce OoC technology in general and demonstrate its power as a tool for infection modelling.
Human in vitro models with true physiological complexity
Organs-on-chip (OoC) represent a breakthrough in understanding host-pathogen interactions and evaluating antimicrobial treatments. The innovative approach to simulate human organ biology on a small-scale level can be used at all stages of drug development, from basic research on molecular mechanisms to pharmacological evaluation of drugs in preclinical research. Such models aid in improving translatability of test results in humans as well as reducing animal testing. Additionally, complex OoC models are capable of including not just pathogens but also a host’s microbiome, immune cells, vasculature, and they can have a multicellular tissue composition.
The underlying concept is based on biochips with cell culture chambers, biocompatible artificial membranes, serving as a scaffold to arrange human cells and providing them structural support, and a variety of channel geometries. These channels are used to introduce the human organ cells into the individual cell culture chamber of each cavity on a biochip, to arrange, combine and separate them in a in vivo-like manner and to continuously supply them with nutrients (Figure 1). They also provide the framework which realizes in vivo-like flow patterns similar to human blood flow.
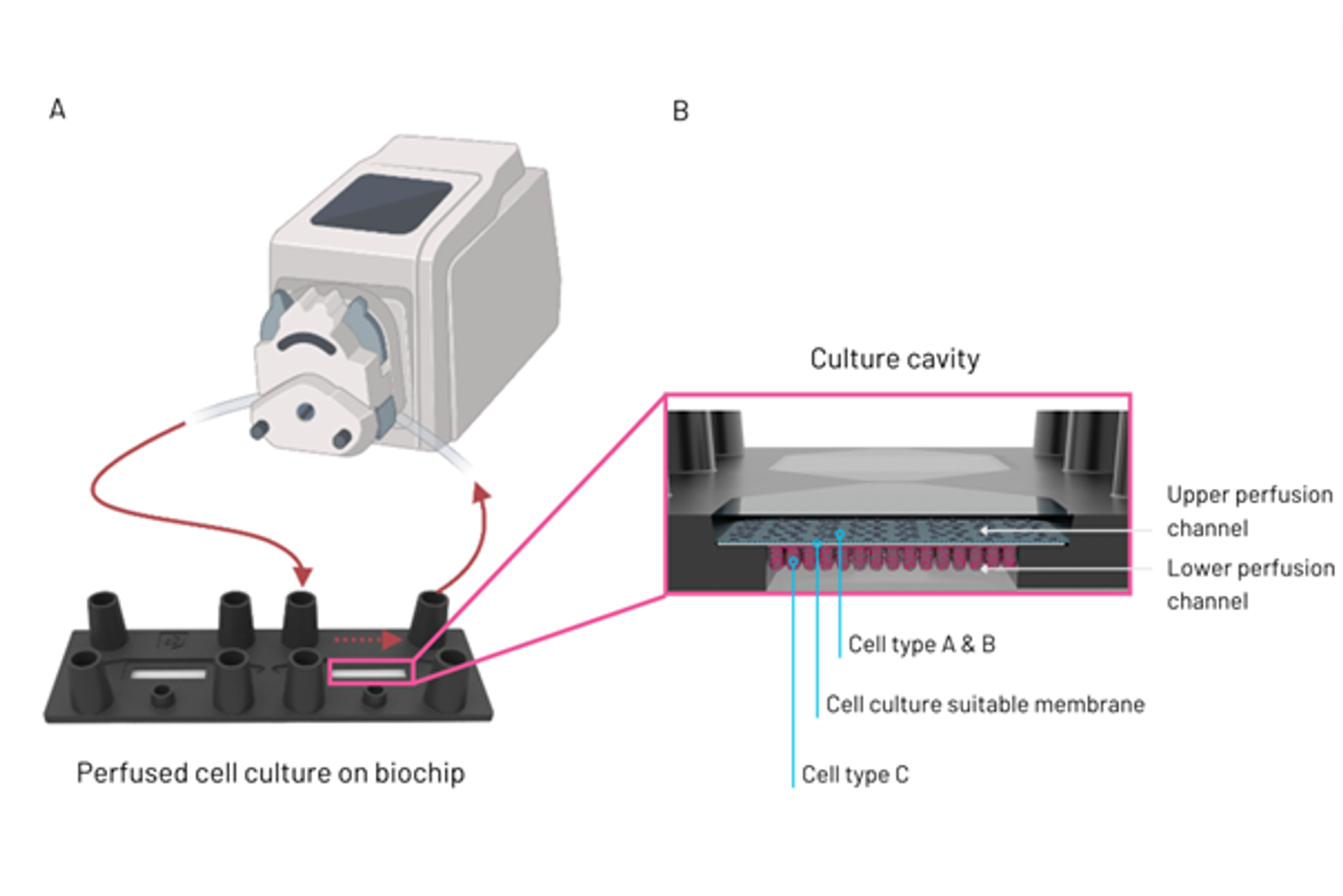
Figure 1: Perfusion set-up of organ-on-chip model, detailing tubing connecting a pump to one of the biochip culture cavities (A). Illustration of culture cavity with upper and lower perfusion channel, including cell types A to C and a cell culture suitable membrane separating the 2 cell culture channels (B).
Complex in vitro models for complex biological processes
Since OoC technology offers enormous flexibility and can be adapted to the various needs of scientific studies, it is highly suitable for investigating intricate processes like infections. The most accurate data can be obtained by using highly complex models that not only integrate all relevant cell types (including immune cells), but also enable the formation of blood vessels to simulate blood flow and include microorganisms such as a host’s microbiome or pathogens.
The basis for the initially mentioned investigation dedicated to studying SARS-CoV-2 disease was an alveolus-on-chip model originally developed by scientist from the University hospital Jena and Dynamic42². This alveolus-on-chip model simulates the architecture in the human lung, complete with vascular and epithelial cell structures co-cultured with macrophages. The model exhibited high barrier integrity, which was further enhanced by physiological flow conditions and the presence of macrophages.
Deinhardt-Emmer et al. used this model to study the complex interplay between pathogens and hosts in pneumonia, one of the most common causes of death worldwide²,³. Influenza virus and Staphylococcus aureus infections triggered a robust inflammatory response in the epithelial cells, which spread to the endothelium and resulted in significant endothelial cell damage, although the integrity of the epithelium remained intact.
In response to the dramatically increased need to understand the process of viral invasion in the context of the COVID-19 pandemic, Deinhardt-Emmer 2021 modified the alveolus-on-chip model by using SARS-CoV-2 permissive epithelial (Calu-3 cells) and vascular cells co-cultured with macrophages¹. It was shown that while SARS-CoV-2 efficiently infects epithelial cells and induces a high viral load as well as an inflammatory response, neighboring endothelial cells are not infected and do not release interferons. However, persistent infection damages both cell types, impairs barrier function and facilitates viral spread, suggesting a complex interplay between epithelial and endothelial cells during SARS-CoV-2 infection.
Understanding the interactions between life-threatening fungi and their host
Infection models based on OoC open completely new possibilities for both basic research and drug development. The latter is extremely important, as resistance rates in widespread bacterial pathogens are alarming worldwide, as the 'Global Antimicrobial Resistance and Use Surveillance System (GLASS) 2022' report shows⁴. The WHO emphasizes that drug-resistant fungal infections are on the rise and because fungal diseases often interact with other infections (e.g. HIV), they are a particular challenge. For example, the emergence and spread of Candida auris, an invasive fungal infection that is resistant to several drugs, is of major concern.
One organism whose interaction with the host can be studied very well in OoC is another member of the genus Candida, Candida albicans, which colonizes the skin and mucous membranes, including the human intestine, without causing symptoms. In certain circumstances, e.g. in immunosuppressed patients, it can lead to dangerous infections that can be life-threatening. Graf et al. (2019) describe an in vitro intestinal model that considers the commensal character of the fungus and its behavior in its natural environment, while conventional models focus exclusively on the pathogenic features of C. albicans⁵.
By including enterocytes as an epithelial barrier and goblet cells to limit fungal adhesion and invasion, significant protection against fungal damage can be demonstrated and it can be shown that the introduction of antagonistic lactobacilli limits the pathogenicity of C. albicans in a time-, dose- and species-dependent manner. In addition, the bacterially controlled shedding of fungal hyphae was identified as a new mechanism for protection against damage.
Going one step further, Maurer et al. (2019) presented another OoC model characterized by the fact that it can be seeded with living microorganisms⁶. In addition, it not only contains tissue-resident innate immune cells, but also exhibits features that are critical for maintaining physiological immune tolerance to microbial-associated molecular patterns. Thus, it represents a unique platform for the study of microbial interactions in a physiologically relevant environment.
If microbiological examination methods are supplemented with image-based analyses, it is even possible to quantify infection processes such as invasiveness and fungal translocation through the epithelial barrier. Recent results (Figure 2) show that C. albicans microcolonies injure the epithelial tissue and cause inflammation⁷. Since macrophages have been integrated into the system, an inflammatory response can also be simulated. In this context, it was shown that the treatment with caspofungin, an antimycotic from the echinocandins group, leads to a reduction in fungal biomass and a change in microcolony morphology, although the efficacy varied depending on the strain used.
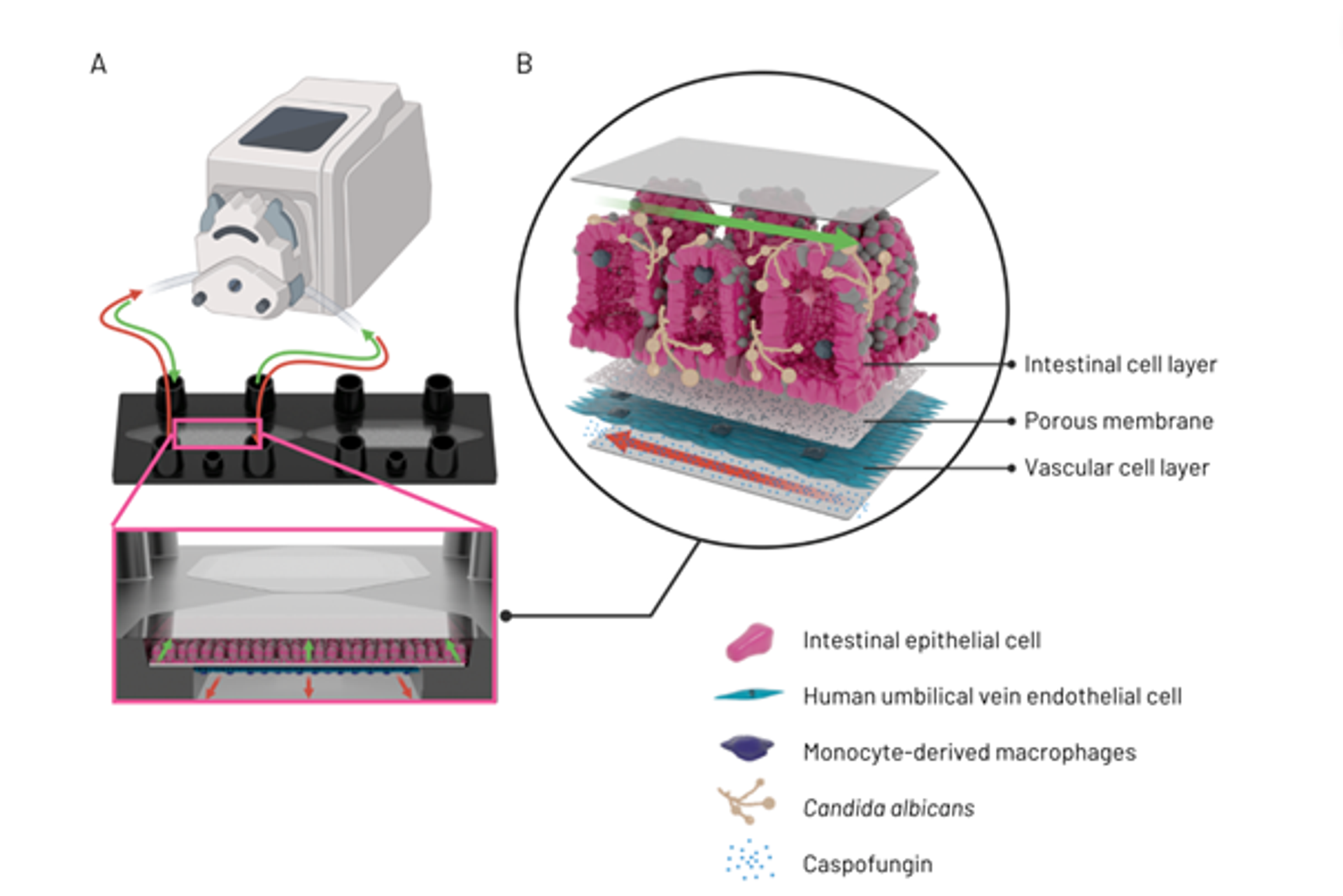
Figure 2: Perfusion set-up of intestine-on-chip model, detailing tubing connecting a pump to one of the biochip culture cavities and a schematic of the flow direction in the upper and lower cavity chamber (A). Illustration of invasive candidiasis in an intestine-on-chip model, detailing vascular compartment with intravenous application of caspofungin, the porous membrane and the intestinal cell layer with Candida albicans hyphae (B).
A growing need for infection models
Despite the outstanding successes in combating infections through improved hygiene, vaccination and antimicrobial therapies, these diseases remain a significant problem in modern medicine. As some chronic neurological disorders that were previously considered degenerative diseases have proven to be caused by pathogens, infectious diseases will continue to be one of the biggest medical problems in the future⁸.
Some even speak of a new era of infectious diseases, characterized by outbreaks of emerging, resurgent, or endemic pathogens, their spread facilitated by globalization and climate change⁹. The importance of infectious diseases and the need for a detailed understanding of the underlying processes will undoubtedly continue to grow. As described, immunocompetent OoC could be a real game changer in the fight against infectious diseases, as they can be used to gain a better understanding of the development of resistance, as well as for designing and validating alternative treatment methods.
This guest editorial article was written and provided by Dynamic42.
References
1) Deinhardt-Emmer, S. et al, J Virol. 2021 Apr 26;95(10):e00110-21. doi: 10.1128/JVI.00110-21.
2) Deinhardt-Emmer, S. et al, Biofabrication. 2020 Feb 19;12(2):025012. doi: 10.1088/1758-5090/ab7073.
3) https://ourworldindata.org/pneumonia
4) https://www.who.int/publications/i/item/9789240062702
5) Graf, K. et al, Dis Model Mech. 2019 Sep 12;12(9):dmm039719. doi: 10.1242/dmm.039719
6) Maurer, M. et al, Biomaterials. 2019 Nov:220:119396. doi: 10.1016/j.biomaterials.2019.119396.
7) Kaden, T. et al, Biomaterials, Volume 307, June 2024, 122525, https://doi.org/10.1016/j.biomaterials.2024.122525
8) Couch, R.B.: Introduction to Infectious Diseases in: Baron S, editor. Medical Microbiology. ©1996, The University of Texas Medical Branch at Galveston.
9) Baker R.E. et al. Nat Rev Microbiol. 2022 Apr;20(4):193-205. doi: 10.1038/s41579-021-00639-z.
