Quantitative microscopy for precision spatial biology: “From samples to knowledge”
In this guest editorial by Dr. Zbigniew Mikulski from the La Jolla Institute for Immunology, explore the fascinating advancements in microscopy and their role in facilitating the generation of high-resolution quantitative data, suited for translational and clinical spatial analysis
14 Sept 2023
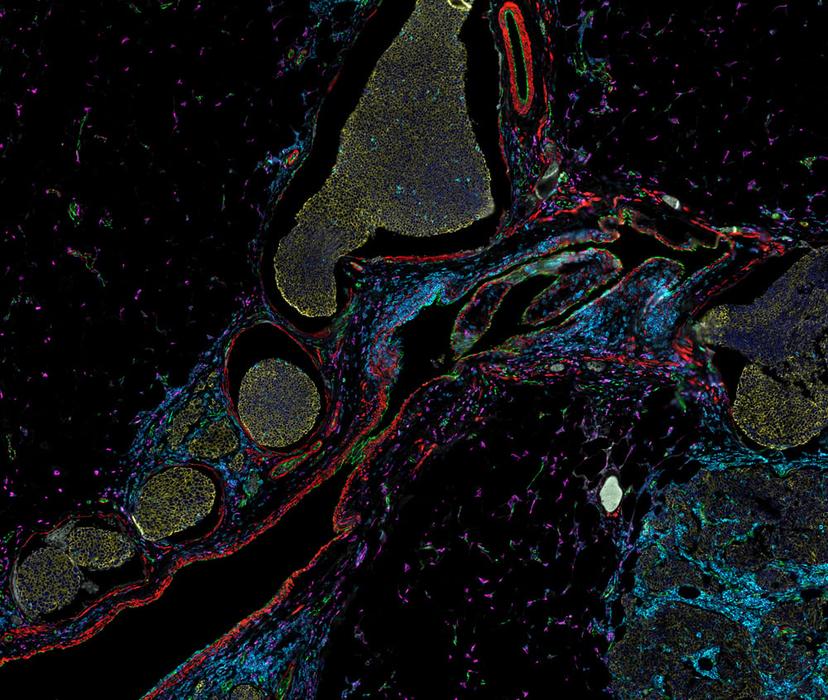
Dr. Zbigniew Mikulski outlines the dynamic evolution of microscopy, highlighting its capacity to provide high-resolution quantitative data tailored for translational and clinical spatial analysis. Dr. Mikulski explains the features required by today’s cutting-edge technologies to support robust, multi-modal data generation and considerations for experimental design and execution to ensure reliable, reproducible results.
Dr. Zbigniew Mikulski is the Director of Microscopy at the La Jolla Institute for Immunology (LJI) and is a globally recognized expert and instructor in the use of spatial biology platforms. His team utilizes multiple types of fluorescence and brightfield microscopy to understand the biology underlying cellular structure, functions, and interactions in health and disease. In this guest editorial, he describes some key requirements to generate trustworthy quantitative data and take spatial biology from biomarker discovery to the clinic.
Removing bias – whole-slide analysis reveals the whole picture
Tissue microarray slides (TMAs) are often used in spatial analysis as a surrogate for whole-slide patient biopsies. Rather than investigating an entire tissue section, small regions are selected, punched out, and assembled into an array, allowing multiple samples to be analyzed at once. While popular, this approach is primarily a compensatory measure to make up for throughput limitations of historically available spatial biology platforms. Such platforms were either only capable of analyzing small regions of interest (ROIs) or had workflows too slow to support the workload of a typical imaging core.
It has been demonstrated, however, that accurate assessment of disease-relevant structures requires the statistical power of whole-slide imaging and removing the bias of small TMA regions1. The LJI team relies on whole-slide analysis for valuable disease insights. Robust measurements can provide definitive evidence for the role of molecules in pathogenesis, such as the degree of expression of antigen-presenting proteins across pancreatic islets of patients with Type 1 Diabetes (T1D)2. “Being able to analyze every single islet is important because when you are working with a disease that is as heterogeneous as T1D, you don’t want to make a clinical decision based on a small subset of islets. It is important to be able to capture large regions of the slide,” explains Dr. Mikulski.
Resolution – why it is important and how it is achieved
Observing the expression of protein biomarkers – singly or in combination – is useful to classify cell types and tissue microenvironments. However, just seeing that a cell expresses a particular protein is not enough. Dr. Mikulski shares, “We need to be able to determine the compartments within the cell in which the protein is expressed to unlock biological significance. Subcellular resolution is required to see critical cell-cell surface interactions and the shapes and number of critical organelles.” LJI investigators studying colitis have demonstrated that the shape and number of mitochondria in neutrophils, and the expression levels of biomarkers within these organelles, correlate directly with disease pathogenesis3.
The resolving power of a spatial imaging system is determined via a combination of two main features – the numerical aperture of the objective and the pixel size of the camera used for detection. Desirable platforms employ high NA objectives to collect maximum photons and reveal subcellular structure while maintaining an appropriate depth of field to accommodate thickness – or sample-prep derived – irregularities in tissue samples. The pixel size, or bit-depth, of the detection camera, contributes to both the image resolution and the dynamic range of signal intensity measured.
Quantitation and the importance of a broad dynamic range
Not only do investigators need to determine which biomarkers are expressed, and their subcellular location, but accurate quantitation of the amount of protein expressed – and differences in expression levels between samples – are critical to generating useful translational data. As described in a recent interview with Dr. Arutha Kulasinghe from the University of Queensland4, of all the ‘Omic’ methods used in spatial biology approaches, proteomics serves as perhaps the most vital because it is the closest assay to the clinic.
Dr. Mikulski explains that rather than just providing cell type identification via a yes/no answer for whether a protein is expressed in a cell, new platforms make it possible to measure varying levels of protein accurately - including very low- and high-expressed proteins that are co-located in a cell. This is accomplished through experimental design to avoid saturation of signal at the high end and using a platform with a highly discerning camera that can differentiate orders of magnitude in signal range. Fluorescence imaging is best suited for that task, especially when paired with strategies to suppress or remove endogenous autofluorescence. Dr. Mikulski cites the work of Dr. David Rimm at Yale, where levels of biomarker expression are quantitated in a highly reproducible manner and used to stratify patients as candidates for advanced therapies5. Quantitative immunofluorescence facilitates ‘measuring versus reading’ 6 and holds promise for transforming the field of quantitative pathology7.
The ideal spatial biology platform

When asked “What makes an ideal spatial biomarker platform?”, Dr. Mikulski recommends an instrument that provides a broad dynamic range of biomarker quantitation in a highly reproducible way, while also delivering subcellular resolution across an entire sample slide.
To achieve a high dynamic range, he recommends fluorescence imaging platforms with scientific cameras that discern many degrees of signal intensity. An 8-bit camera delivers a maximum of 256 gradations whereas a 16-bit camera will discern 65,535 gradations of intensity across the same image. “High dynamic range is extremely helpful when working with biological samples as you often don’t know how much signal to expect. The imaging conditions should be set for the highest likely intensity to avoid saturation, and powerful spatial biology platforms will allow accurate quantitation of both high- and low-abundance markers in the same analysis.”
The ability to generate both biomarker quantitation and H&E images from the same slide is highly desirable since this multi-mode analysis provides both molecular information and morphological context – thus delivering significant insight into the disease state of the tissue. It also helps to engage with the pathologists who mostly see tissues through the colors of the classical H&E stain. “The ability to easily access the collective knowledge of pathologists is an incredible strength,” says Dr. Mikulski. He advocates for multidisciplinary teams where the staff doing sample preparation, tissue imaging, computational analysis, and interpretation work together at each step of the project.
Pilot studies are necessary for assay optimization and for executing ‘power analysis’ to determine the number of patient samples that must be analyzed and correlated with clinical outcomes to provide statistical robustness. A fast and straightforward workflow with a one-step stain and scan of many markers simultaneously is the preferred ‘agile’ method versus lengthy workflows that require many serial steps with assay performance revealed only after significant time and reagent cost.
To meet their need for rapid and cost-effective assay development, validation, and implementation, the team at LJI favors the one-pass, highly multiplexed Orion platform, versus cyclic multiplex IF methods that take many days to execute and greatly prolong new assay development and optimization.
Dr. Mikulski will host a QuPath Training Course: From Samples to Knowledge at the La Jolla Institute for Immunology on October 9–11, 2023. This introductory course will be followed by a Multiplex Image Analysis workshop organized in partnership with the Quantitative BioImaging Society on October 12–13, 2023, also at the La Jolla Institute.
RareCyte will be in attendance and featuring the Orion platform. Orion brings analytical rigor to spatial analysis in a timeframe that makes it applicable for the validation and clinical utility of spatial biomarkers.
References
Lin, J.-R. et al. Multiplexed 3D atlas of state transitions and immune interactions in colorectal cancer. Cell. 2023 Jan 19;186(2):363-381.e19. DOI: 10.1016/j.cell.2022.12.028. Multiplexed 3D atlas of state transitions and immune interaction in colorectal cancer - PubMed (nih.gov)
Quesada-Masachs, E. et al. Upregulation of HLA class II in pancreatic beta cells from organ donors with type 1 diabetes. Diabetologia 2021 DOI:10.1007/s00125-021-05619-9 Upregulation of HLA class II in pancreatic beta cells from organ donors with type 1 diabetes - PubMed (nih.gov)
Riffelmacher, T. et al. Metabolic activation and colitis pathogenesis is prevented by lymphotoxin β receptor expression in neutrophils. Mucosal Immunol 2021 DOI 10.1038/s41385-021-00378-7 Metabolic activation and colitis pathogenesis is prevented by lymphotoxin β receptor expression in neutrophils - PubMed (nih.gov)
Kulashinghe, A. Bringing spatial biology to the clinic: “A new lens on cancer biology.” Editorial article, Select Science. October 2022 Bringing spatial biology to the clinic (selectscience.net)
Martinez-Morilla S. et al. Quantitative assessment of PD-L1 as an analyte in immunohistochemistry diagnostic assays using a standardized cell line tissue microarray. Laboratory Investigation 100: 1 January 2020 https://doi.org/10.1038/s41374-019-0295-9 Quantitative assessment of PD-L1 as an analyte in immunohistochemistry diagnostic assays using a standardized cell line tissue microarray (nature.com)
Rimm, D. Precision Medicine vs Persuasion Medicine: Measuring vs Reading HER2. Yale Cancer Center Grand Rounds | February 21, 2023 Yale Cancer Center Grand Rounds | February 21, 2023 - YouTube
Rimm, D. Measuring versus reading - transforming the field of quantitative pathology. Editorial article, Select Science. February 2023 Measuring vs reading: Transforming quantitative pathology (selectscience.net)

