Recombinant protein purification process development
A case study comparing process development for the purification of recombinant proteins using Cytiva Protein Select resin vs a nonaffinity method
17 Dec 2024
Therapeutic proteins must be produced with a very high level of purity, but proteins other than mAbs often don't have an affinity binding partner to facilitate their purification. To be able to standardize and simplify the purification with an affinity step, one strategy has been to add a tag to the recombinant protein. Historically, using a tag has mainly been used for research purposes and not for biotherapeutic purification because tag removal was not traceless, and could generate modifications of the target protein. With Cytiva™ Protein Select™ technology, process development can be standardized with the use of a traceless and self-cleaving tag as described in this animation. This shortens the overall development time while maintaining — or improving — product quality in fewer steps.
In this study, we compare an affinity purification-based process using the Cytiva™ Protein Select™ resin and tag technology to a conventional approach (referred to as the "non-affinity process") for purifying a recombinant protein. Our protein of interest, in this case, was a thermostable scaffold protein expressed in E. coli. This protein is used as an antibody mimetic that binds to a desired ligand and is selected from a library of engineered variants. We present the results of screening and transfer to columns suitable for scale-up studies, comparing development time, recovery and purity of the protein of interest between the non-affinity method and the affinity method using Cytiva™ Protein Select™ technology.
Part 1: Screening
We first performed screening in small scale (using HiTrap™ chromatography columns) to identify the techniques and resins to carry forward to larger scale. The requirements for each process were at least 95% purity and at least 50% recovery of the protein of interest.
Non-affinity process
We ran 49 experiments testing various combinations (buffers, techniques, and resins) to identify a protocol for purification of the scaffold protein without the use of Cytiva™ Protein Select™ technology. This screening process took 11 weeks and resulted in choosing a three-step protocol starting with anion exchange chromatography (AIEX) for the capture step, following by a heat treatment, and hydrophobic interaction chromatography (HIC) as a polishing step.
The protein purity obtained with this protocol was 95%, as measured by analytical size-exclusion chromatography (SEC) at 214 nm (see Fig 1), which was our goal for the screening. However, it's worth noting that for production of therapeutic molecules, purity of 95% would be too low, thus requiring either more optimization later in development or addition of an extra polishing step before a final process optimization. Either scenario would require additional development time.
Affinity process using Cytiva™ Protein Select™ resin
We ran 17 experiments to identify a suitable protocol based on Cytiva™ Protein Select™ technology, and this required 3 weeks of development time. We selected a two-step protocol consisting of an affinity capture step with Cytiva™ Protein Select™ resin and an AIEX polishing step, which delivered a purity level close to 100% according to analytical SEC (Fig 1). We also analyzed the purified protein by LC-MS to confirm a traceless removal of the tag (Fig 2). The analysis showed that the scaffold protein purified with Cytiva™ Protein Select™ technology had a mass of 16 876.7 Da, which is the mass expected after tag removal, confirming that the protein was pure and in native state with no traces of the tag amino acids.
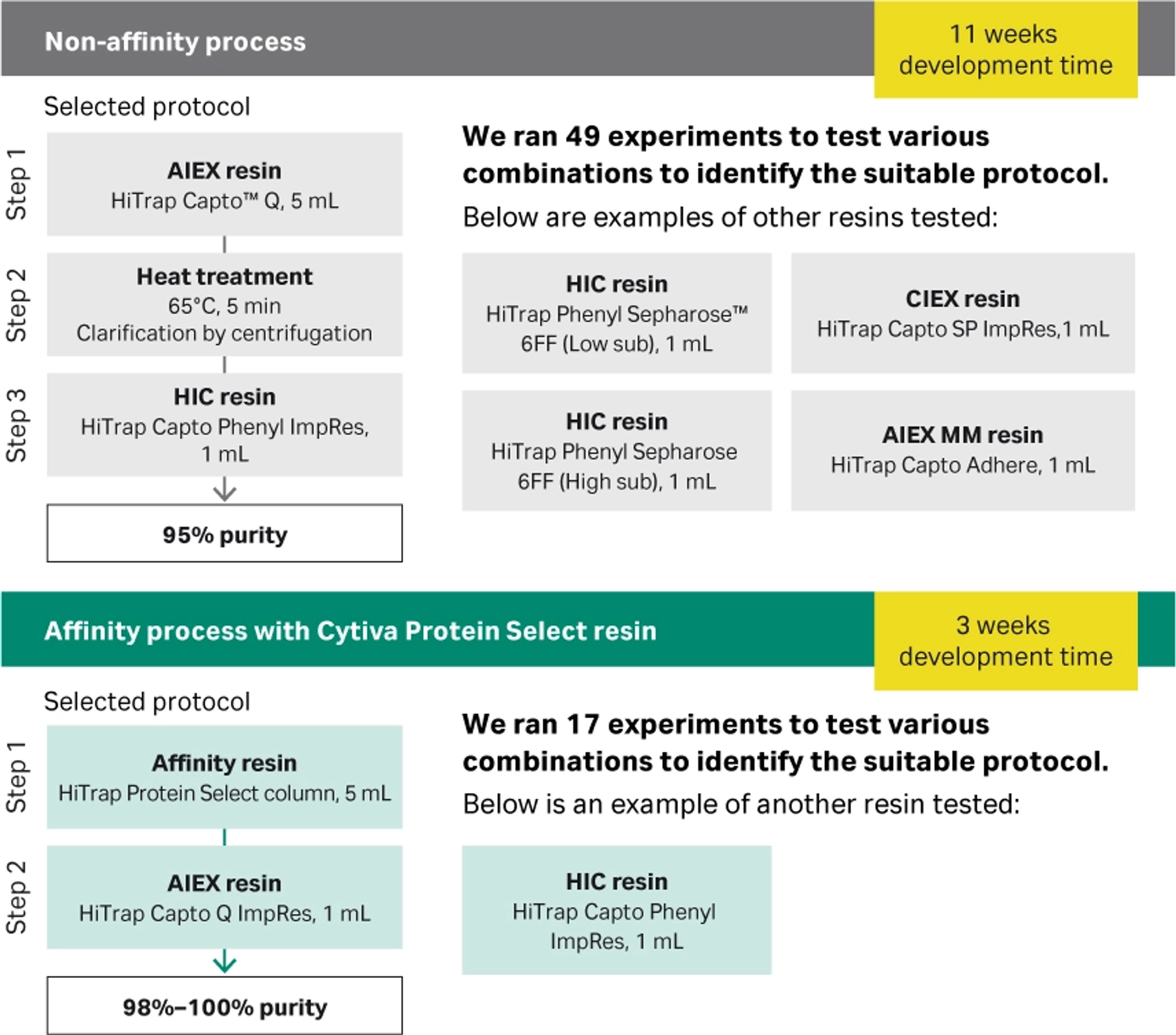
Fig 1. Development of the small-scale affinity-based process using Cytiva™ Protein Select™ resin took less time (3 weeks vs 11 weeks for non-affinity process) and was less complex (2 steps vs 3 steps). AIEX = anion exchange chromatography; CIEX = cation exchange chromatography; MM = mixed mode chromatography; HIC = hydrophobic interaction chromatography
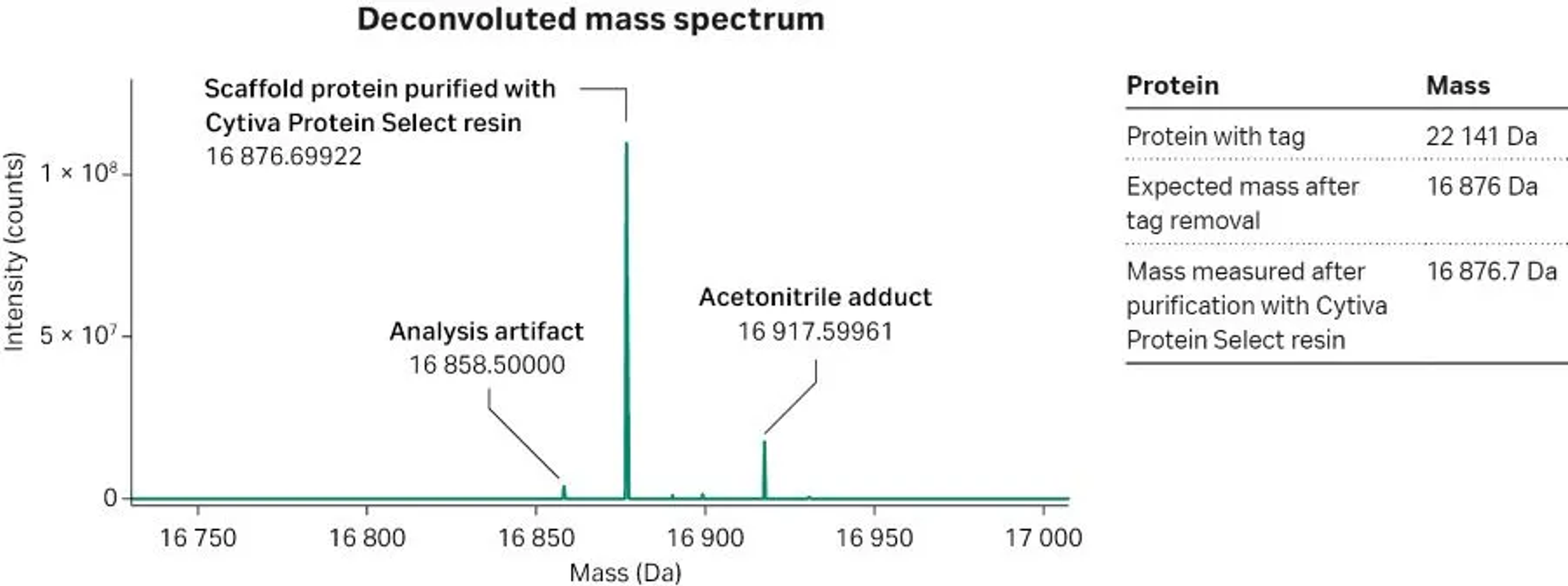
Fig 2. Mass analysis of the purified protein after tag cleavage. Mass error = 40 ppm. LC-MS analysis on Waters ACQUITY, C8-RPC.
Part 2: Process performance at 10 cm bed height
The next step was to test the protocols we selected in Part 1 at larger scale — in this case, in 10 cm bed-height chromatography columns — and to compare the processes' performance in terms of purity and recovery.
Note that to facilitate recovery analysis of full-length fusion protein in crude cell extracts, we added a 6×His epitope tag upstream of the Cytiva™ Protein Select™ tag. We used the same protein construct for both processes to enable a fair comparison (Fig 3).
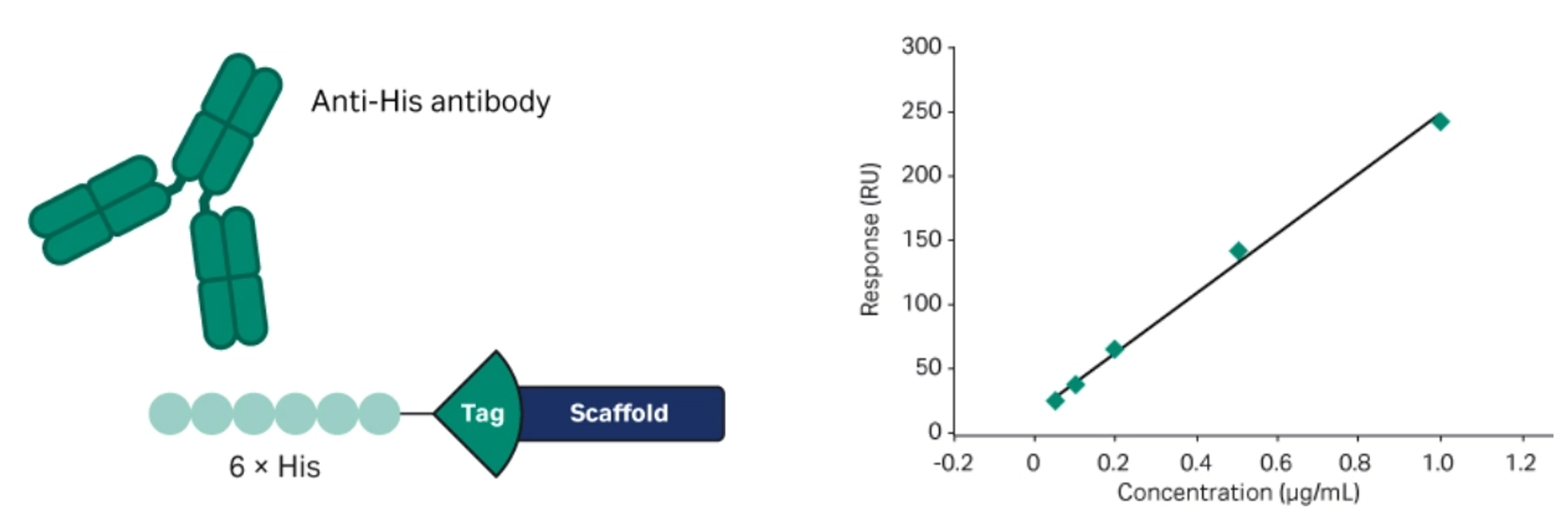
Fig 3. (Left) Protein construct with upstream 6×His tag allowing Biacore™ SPR analysis using anti-His antibodies. (Right) Calibration curve using purified fusion protein as standard for recovery calculations.
Results of performance testing
At this larger scale, we obtained a purity of 91-92% with the non-affinity process, vs 98% with the affinity process using Cytiva™ Protein Select™ technology. Recovery with the non-affinity process was 57%, vs 53% with the affinity process (Figs 4 and 5).
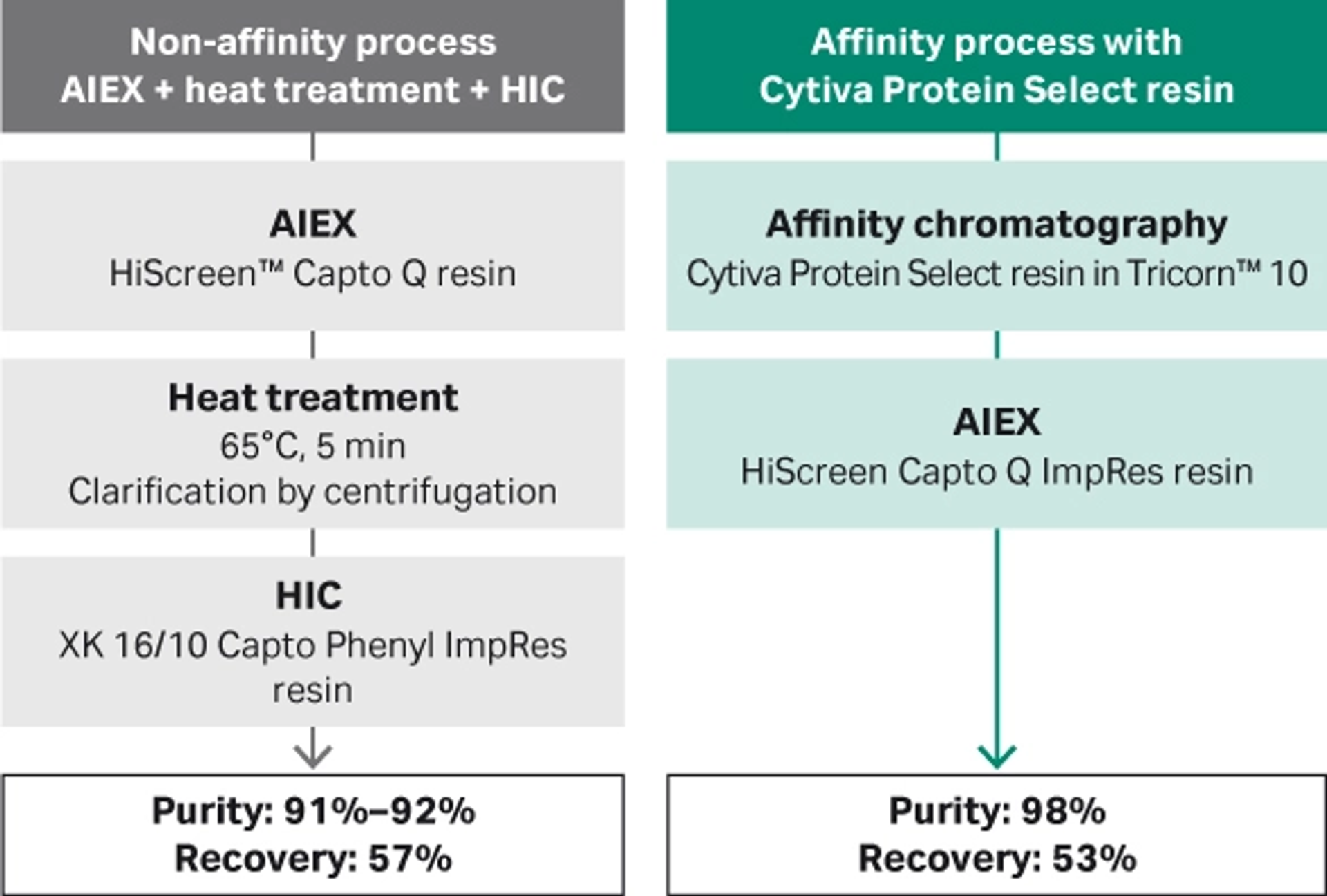
Fig 4. Comparison of process performance at 10 cm bed height.
Figure 5 shows the results from analytical SEC. The different retention times reflect the uncleaved or cleaved scaffold fusion obtained in the processes.
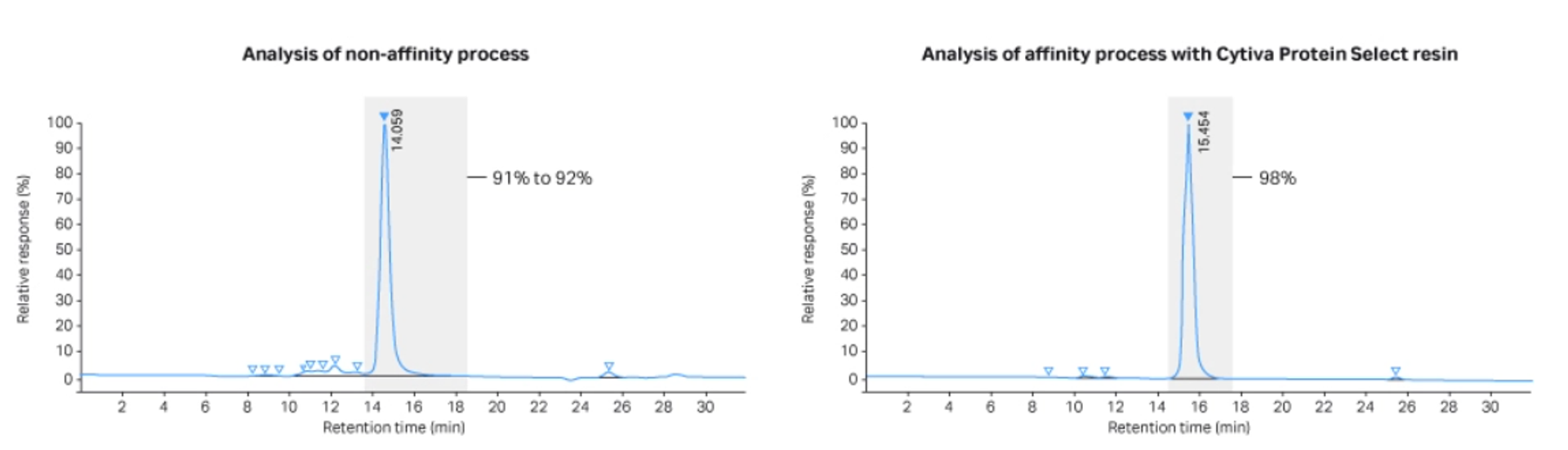
Fig 5. Analytical size exclusion at 214 nm on a Superdex™ 75 Increase 10/300 GL column. The non-affinity process (Left) resulted in a protein purity of 91% to 92%, whereas the affinity purification method using Cytiva™ Protein Select™ resin (Right) resulted in protein purity of 98%.
For the non-affinity process, a low absorption coefficient at 280 nm made pooling criteria difficult in the first step (Fig 6, left). Therefore, extensive analytical work was necessary to identify the fractions to be pooled for the next step. In contrast, the pool from the affinity purification step (Fig 6, right) was much easier to identify, which reduced the amount of analytical work required.
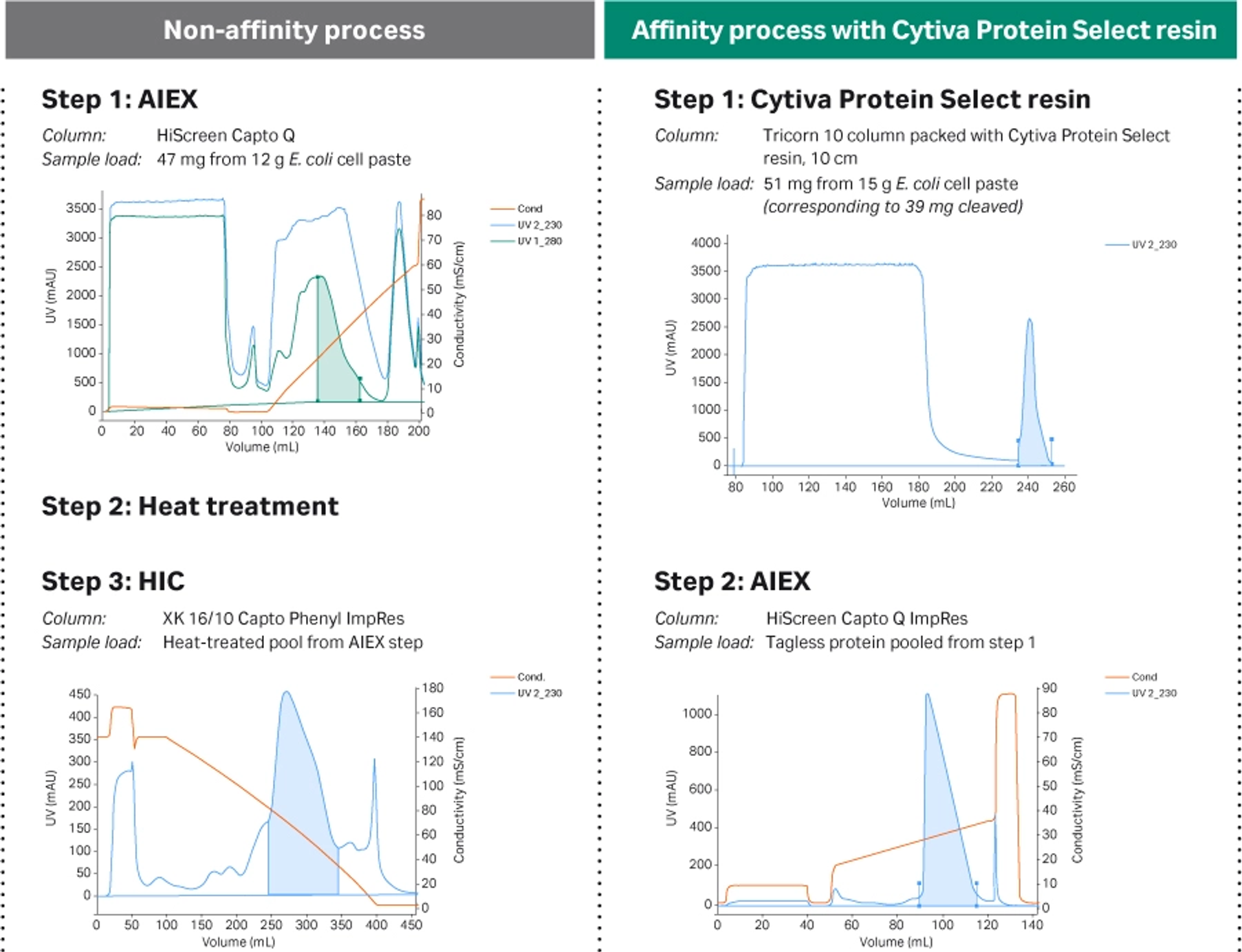
Fig 6. Chromatograms obtained from the different processes. The shaded areas correspond to the pooled fractions.
Conclusion
In this study we compared Cytiva™ Protein Select™ technology for purification of a recombinant protein that does not have an affinity binding partner to conventional methods requiring more labor-intensive process development and more polishing steps to achieve purity suitable for biotherapeutic applications. Our results, summarized in Table 1, show that this system enables predictable transfer to larger bed heights, faster method development, and higher-purity protein in fewer steps. By simplifying process development time and reducing the number of purification steps, this technology is a valuable tool for increasing throughput in labs developing candidate molecules for preclinical studies.
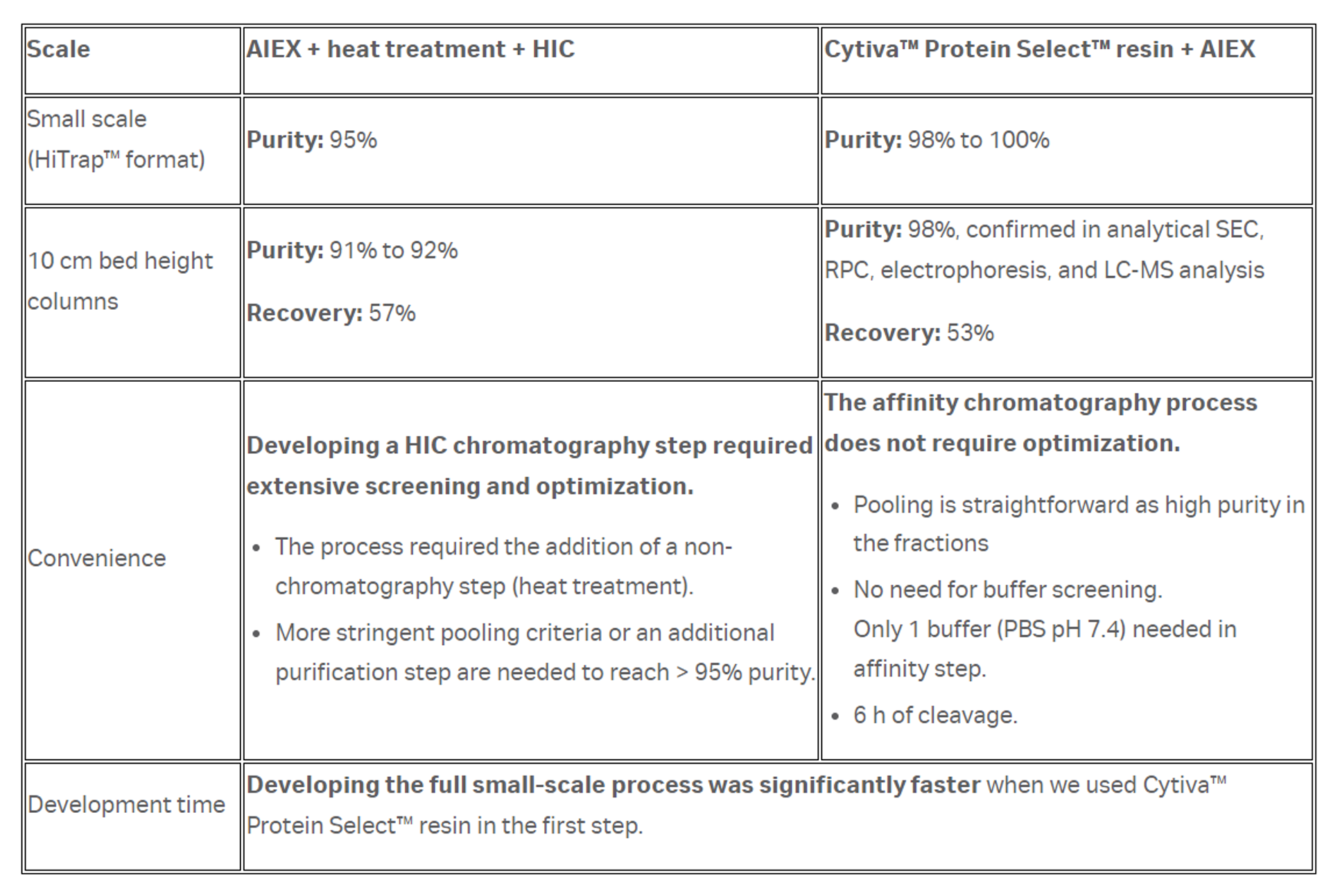
Table 1. Recap of comparison of both methods
*This article was authored by Johan Ohman, Cytiva.

