The Indispensable Need for High-Quality Nucleic Acids in Next-Gen Sequencing
Josh Haimes, technical lead at ArcherDX, explains how successful NGS is influenced by step-by-step quality control
8 Jan 2018

SS: Briefly describe your role at ArcherDX.
JH: I am the technical lead for the applications group and the production sequencing group at ArcherDX. The applications group administers Archer’s NGS platforms to contrived and clinical sample types to understand and demonstrate assay performance and explores the bounds of new NGS applications. The production sequencing group, on the other hand, is engaged in collaboration activities and screening contracts. Efforts from both the groups often result in peer-reviewed publications, application notes or marketing collateral.
How did you become interested in NGS?
My first project employing NGS was in 2011. Our goal was to improve the reproducibility of pooled shRNA screens. We performed viability screens with a library of ~10,000 shRNAs and determined reproducibility with NGS analyses. The results are published in Strezoska et al., PLoS One, 2012.
What project are you currently working on?
My groups are currently working on several projects and collaborations. Some of the applications include immune cell receptor sequencing, characterizing novel genetic fusions in sarcoma, CAR-T engineering, liquid biopsy research, and companion diagnostics.
Are there projects where you use FFPE samples?
Yes, FFPE is a common sample type for input into Archer’s NGS workflows. We see FFPE more commonly than fresh-frozen sample types. Cancer diagnosis has relied heavily on morphological characterization such as H&E staining and immunohistochemistry (IHC). Both approaches require a fixed, intact sample, hence the FFPE method of tissue preservation has been traditional. Precision molecular oncology, however, is still in its adolescent stages. As H&E and IHC continue to play a valuable role in cancer diagnostics, FFPE is still the prevailing source of nucleic acids for cancer omics research. It’s unfortunate, given the impact of formalin fixation on nucleic acid suitability for downstream enzymatic manipulation, but we must make the most of this readily available tissue.
What are some challenges you’ve faced when extracting nucleic acids from FFPE samples?
The main challenge with FFPE is the variability in the quality of the resulting nucleic acid. For example, the amount of information that can be recovered from 10ng of nucleic acid from FFPE is not the same from sample to sample. Without proper QC checks, inaccurate assumptions about assay performance or even false negative results can occur.
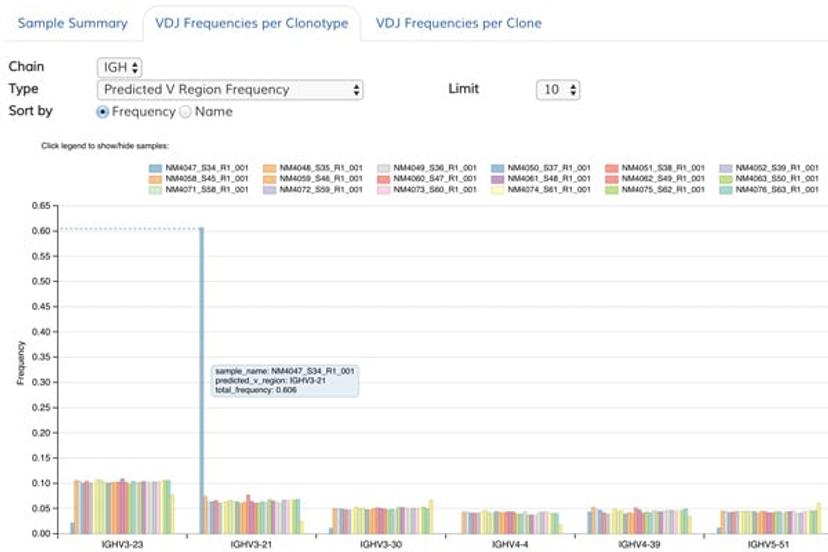
B-cell receptor sequencing detects the clonal nature of the Raji Burkitt’s Lymphoma B-cell line. Image courtesy of ArcherDX.
How did you overcome those challenges?
We have explored nucleic acid purification from FFPE and have published results of our findings in an extraction white paper where we studied the impact of three important steps in nucleic acid extraction from FFPE samples: the mode of paraffin removal from samples, crosslink reversal time and temperature, and finally, the pH for extraction. Beckman Coulter’s FormaPure was one of the first extraction kits we found success with, and we continue to recommend this kit for purification. On occasion, we have found genetic variants in samples purified using FormaPure that we could not otherwise recover using other kits that yielded poor quality nucleic acid. NGS users also find the bead-based format of the FormaPure kit familiar, as it is similar to the AMPure XP clean-ups commonly used in library preparations.
What recommendations would you have for NGS users with FFPE as a starting sample?
Archer has put forth a great deal of effort as a thought leader and educational resource surrounding this topic. Extraction is a critical variable for NGS, and knowledge of the quality of resulting nucleic acid guides NGS library input amount and sets expectations for sensitivity. So we have engineered sample QC into our workflows to position users of Archer NGS for success. The Archer PreSeq DNA and RNA QC Assays inform scientists of the quality of the DNA or RNA in their sample before targeted library prep for NGS, providing a guidance on input. Plus, Archer’s sequencing metric analytics provide a report on NGS library complexity, which indicates target coverage and is correlated to assay sensitivity.
Overall, we feel that the scientific community is much more aware of sample quality today than it was five years ago.
Do you see the applications of FFPE samples in genomics evolving?
It’s likely that FFPE will serve a purpose for structural observations in traditional molecular pathology that benefit from the fixation, for example, immunohistochemistry and fluorescent in situ hybridization. However, as adoption of nucleic acid-based detection methods widens, one might hope that FFPE fades as the primary sample type for assays such as NGS that do not require fixation. Nearly all aspects of NGS assay performance are enhanced with fresh frozen samples versus FFPE, simply because of the increased length and amount of amplifiable molecules present. But until there is a global change in the standard operating procedure of fixing or freezing samples, education around the effects of the sample fixation on nucleic acid quality is paramount to obtain reproducibility.
What are the future projects at ArcherDX?
Archer has developed and optimized chemistries and bioinformatics pipelines for NGS library preparation across a range of applications. These include detection of fusions, single nucleotide variants, indels, copy number variation, expression signatures and more. Recently, we launched products for immune cell receptor sequencing and liquid biopsy research.
Another area of our focus is the user experience and turnaround time for Assay Designer, the online tool that enables users to design custom NGS panels that utilize the power of anchored multiplex PCR. As the field is changing so quickly, it’s becoming more important for labs to be able to easily expand a panel with new targets without impacting performance.
What might be the next milestone for NGS?
Personally, I am excited by immune cell receptor sequencing and immune oncology. I’m also interested in topics surrounding liquid biopsy NGS, circulating free DNA and exosomes.
