Transforming biomarker discovery and diagnostics with highly multiplexed aptamer-based proteomics
How can a proteomics platform that can measure over 7,000 protein biomarkers from one sample be used to assess an individual’s overall health, monitor disease progression, and inform healthcare decisions?
1 Mar 2023
As accessible indicators of disease, proteins present in bodily fluids and tissues have captured the attention of scientists and clinicians alike, leading to extensive research into new predictive, diagnostic, and prognostic biomarkers to inform healthcare decisions. The process of biomarker discovery and validation, like the proteins and their respective biological activity, can be vastly complex, often involving a team of medical, analytical, and bioinformatics collaborators. Such a team however, can only progress as far, and as fast, as the available technology will allow.
The challenges with existing analytical proteomic platforms such as liquid chromatography coupled to mass spectrometry (LC-MS), two-dimensional electrophoresis (2DE), and antibody arrays are that they all have inherent limitations with sample throughput and precision. For example, antibody-based detection of bound proteins is a widely used method of identification and quantification, but the specificity of an antibody to detect a given protein can be poor, and resultant crosstalk difficult or impossible to assess.
A vision of massively multiplexed proteomics
Recognizing the need for a new technology that could measure large-scale proteomics with greater sensitivity, specificity, and reproducibility, Jeff Carter, Senior Director, Chemistry, joined SomaLogic in 2001. He had a vision and mandate to develop synthetic oligonucleotide processes to support a massively multiplexed proteomics platform, leveraging his experience within aptamer-based therapeutics that he gained whilst working for the company’s predecessor, NeXstar Pharmaceuticals. “We felt aptamers were the best way to achieve our vision. With the understanding that they could be used as the structural component leading to affinity binding to proteins, we would then be able to obtain a read out on conventional DNA microarrays or sequencing-based technologies,” explains Carter.
A few years in, Carter and his team realized some significant technical challenges, which ultimately led them to evaluate, and then implement, the addition of unique modified nucleotides. “These modified nucleotides involved taking amino acid side chains and incorporating them into DNA with the simplistic view that DNA is a hydrophilic molecule and doesn't have some of the functionality that proteins have,” he states. “Essentially, we can make DNA look and act more like proteins, which are in turn more likely to bind to proteins.”
The conception of SOMAmer® reagents
In search of the ideal nucleotides, Carter and his team evaluated several different modified bases, fundamentally looking at amino acid side chains, and eventually landing on planar aromatic hydrophobic amino acids. “We found those worked the best, and it was an intuitive step in the sense that proteins often have hydrophobic patches that are encompassed by phenylalanine, tyrosine, and tryptophan, which are overrepresented in many active sites,” states Carter. “We saw great success with incorporating them into our aptamers. Not only a higher success rate with any given protein, but also improved thermodynamics, higher binding affinities, and crucially, slower off rates. This led us to coin the term SOMAmer reagents (slow-off rate modified aptamers).”
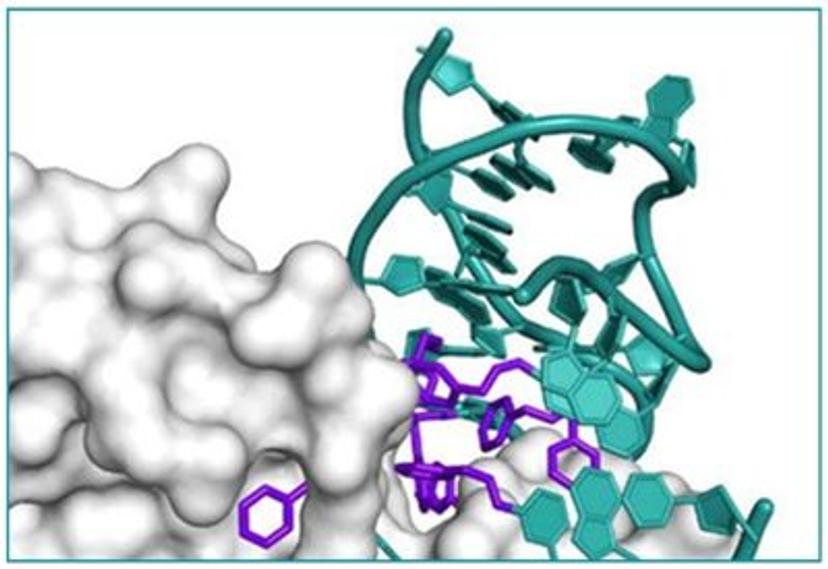
Fast forward to present day, and SomaLogic now have a vast library of 7,000 aptamers to measure 7,000 unique proteins within the proteome, originating from multiple different matrices like plasma, serum, urine, cerebrospinal fluid, and synovial fluid. With the help of numerous publications1-14 and supporting patents14-17, Carter and his team have also now been able to apply these unique reagents to a multiplexed proteomics platform called SomaScan®.
“With SomaScan, the aim is to use this as a large-scale proteomic discovery assay. We're looking for biomarkers of disease states, and so we've run many different clinical trials collecting plasma and serum samples from either research institutes, academic labs, or pharmaceutical companies who were working to identify disease markers for either diagnostic or therapeutic interventions,” explains Carter. “Ultimately, the goal was to get to large discovery plex, identify sets of markers, develop diagnostics, and use these life science tools as a diagnostic platform.”
The advantages of collaboration
For Carter and his team, there were significant challenges to overcome when first developing the modified nucleotides for the SOMAmer reagents, particularly as the synthetic capability was not readily available at the time. Leaning on some initial work developed at NeXstar Pharmaceuticals, and leveraging contract manufacturers and the knowledge of chemists who can make phosphoramidites, they were able to produce the first implementations.
Building relationships with pharmaceutical companies, research groups, and clinics also looking to identify new sets of markers to better understand disease biology was vital. The sharing of both clinical samples and data between SomaLogic and these groups helped to inform the SomaScan platform’s development, whilst also improving clinical development and diagnostic programs for these groups in return.
This collaborative approach was also adopted when working with its suppliers, particularly during the discovery process. “Many elements of our assay technology, bioinformatics analysis, and our clinical development programs are built on tools and techniques that have come of age over the last few decades,” explains Carter. “Without the oligonucleotide synthesis and phosphoramidite chemistry during our discovery phase, we wouldn't be able to make the SOMAmers. Without the right equipment and reagents, many of which we sourced from LGC Biosearch Technologies, we wouldn't be able to do what we do, and we’re working with them, along with other suppliers, on a number of other aspects of oligonucleotide chemistry.”
Putting the technology on the front line
Moving forward, Carter and his team intend to continue to develop the number of proteins they can accommodate with the highly multiplexed SomaScan platform and improving its clinical utility in the process, with the next milestone being its adoption in patient screening.
“Depending on who you ask, the entire medically relevant proteome is around 20,000, not including post-translational modifications and numerous different isoforms. From a clinical perspective, we envision the technology being used in routine longitudinal studies to measure an individual’s overall health status, monitor the progression of many diseases, and inform healthcare decisions,” he shares. “A personal goal is to see our technology being used for the early detection of critical, high mortality rate cancers that are otherwise difficult to detect until patients are symptomatic. Pancreatic cancer for example. That’s my vision for the future.”
References
Gold L., et al. 2010. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE. 5(12).
Rohloff J.C., et al. 2014. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol Ther Nucleic Acids. 3(10), e201.
Gawande, Bharat N, et al. 2017. Selection of DNA aptamers with two modified bases. PNAS Early Edition.
Carlson, Michelle, Carter, Jeffrey D, & Rohloff, John. 2015. Improved preparation of 2 M triethylammonium bicarbonate. Green Chemistry Letters and Reviews. 8:3-4, 37-39.
Rohloff, John C, et al. 2015. Practical Synthesis of Cytidine-5-Carboxamide-Modified Nucleotide Reagents. Nucleosides, Nucleotides, and Nucleic Acids. 34:180-198
Gelinas, Amy D, et al. 2014. Crystal Structure of Interleukin-6 in Complex with a Modified Nucleic Acid Ligand. Journal of Biological Chemistry. 289(12)
Davies, Douglas R, et al. 2012. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. PNAS Early Edition
Gupta, Shashi, et al. 2011. Rapid Histochemistry Using Slow Off-rate Modified Aptamers With Anionic Competition. Appl Immunohistochem Mol Morphol. 2011 Jan 7
Gold, Larry, et al. 2010. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE. 5(12), Dec 7.
Vaught, Jonathan D, et al. 2010. Expanding the Chemistry of DNA for In Vitro Selection. J. AM. CHEM. SOC. 132, 4141–4151
Hill, Kenneth W. et al. 2001. Diels-Alder Bioconjugation of Diene-Modified Oligonucleotides. J. Organic Chem. 66, 5352-5358
Bridonneau, Philippe, et al. 1999. Purification of a highly modified RNA-aptamer: Effect of complete denaturation during chromatography on product recovery and specific activity. J. Chromatography B: Biomedical Sciences and Applications. 726(1-2), 237-247
Vargeese, C et al. 1998. Efficient Activation of phosphoramidites by 4,5-Dicyanoimidazole. Nucleic Acids Research, Feb 1998
Patents (pending)
US 10,221,207 5-position modified pyrimidines and their use
US 9,163,056 B2 5-Position modified pyrimidines and their use
US 9,382,533 Method for generating aptamers with improved off-rates
US 8,975,026 Method for generating aptamers with improved off-rates