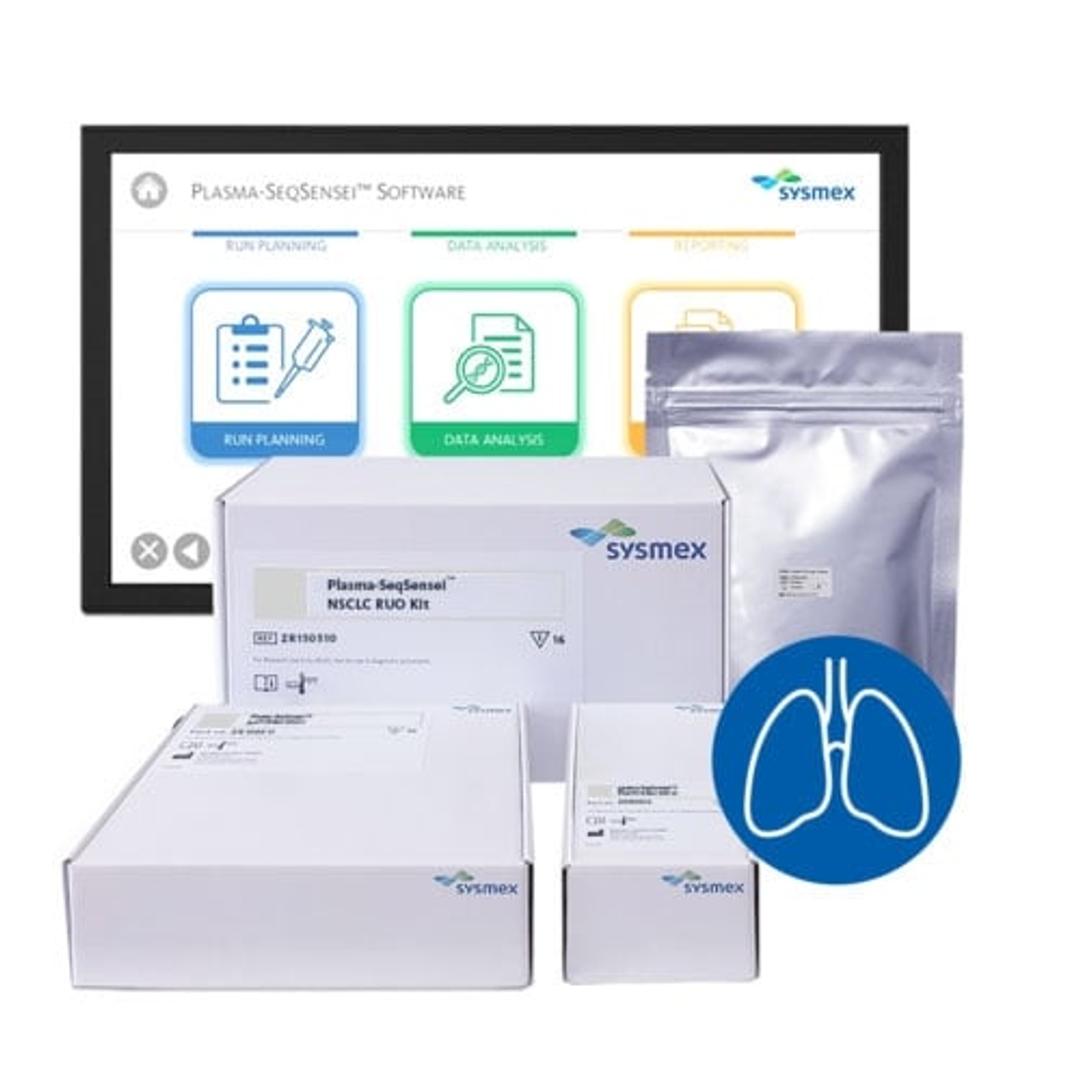
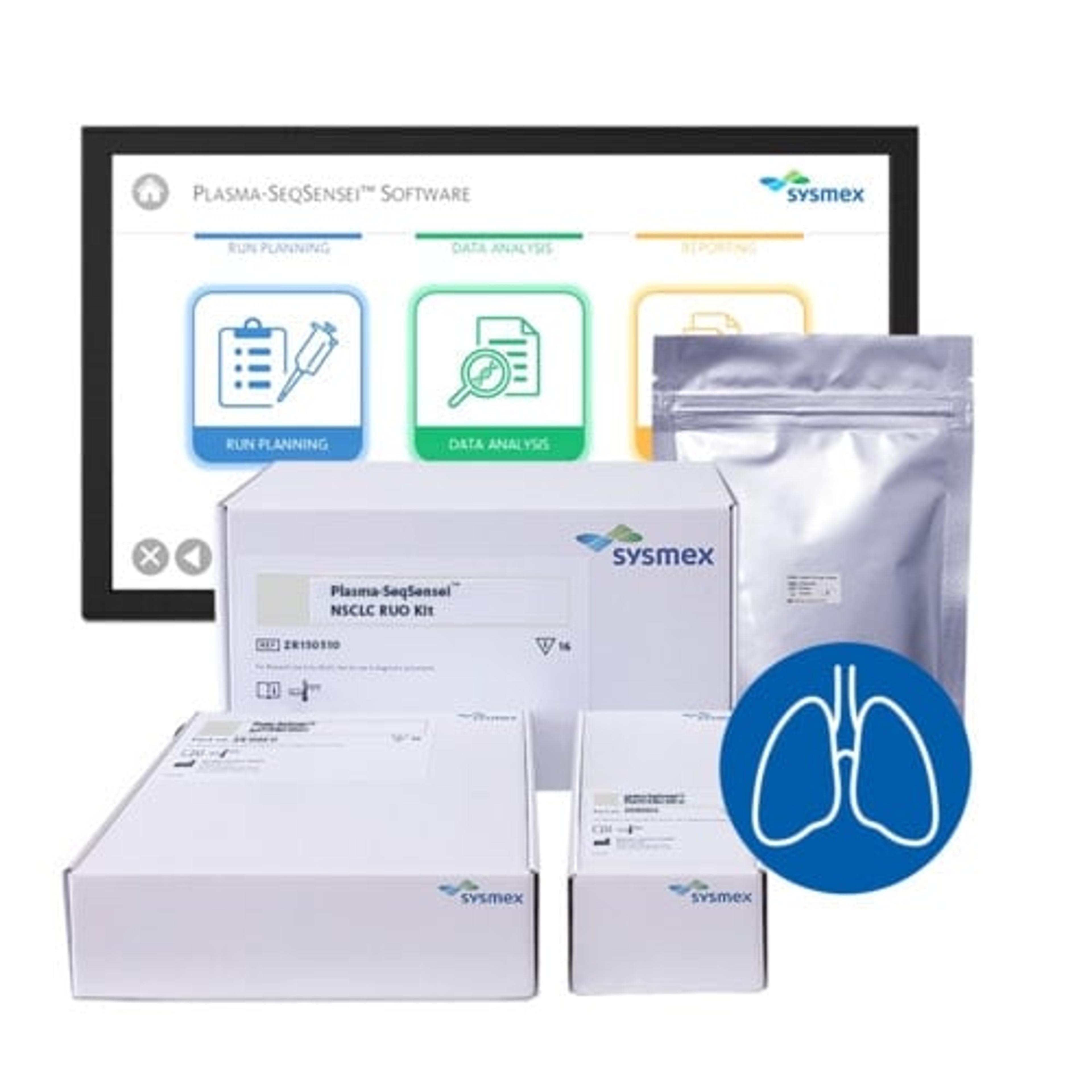
Plasma-SeqSensei™ Non-Small Cell Lung Cancer RUO Kit
The Plasma-SeqSensei™ (PSS) NSCLC RUO (research use only) Kit detects mutations in circulating tumour DNA (ctDNA) from plasma in patients with non-small cell lung cancer.
The Plasma-SeqSensei™ (PSS) NSCLC RUO (research use only) Kit detects mutations in circulating tumour DNA (ctDNA) from plasma in patients with non-small cell lung cancer.
The kit uses next generation sequencing for the detection and identification of mutations in BRAF, EGFR, KRAS, and PIK3CA genes in human cfDNA isolated from blood plasma which play a significant role in the development of NSCLC and are relevant biomarkers for prognosis, choice of therapy as well as monitoring of recurrence and therapy response. [1] [2]
EGFR gene mutations (in exons 18-21) coding for the internal receptor tyrosine kinase [TK] domain of EGFR have a variable ability to activate the TK in the absence of ligand binding. EGFR mutations are reported in 10-15% of Caucasian adenocarcinomas (all cases regardless of smoking history) and in 40–60% of adenocarcinomas in East Asian populations. [3]
BRAF mutations are the driver oncogenes in 1-3% of cases of NSCLC (classic V600E form (50%)). [4]
KRAS mutations occur in ~30% of lung adenocarcinomas (KRAS G12C comprises ~44% of all KRAS mutations, so finally making up mutations in ~13% of all lung adenocarcinomas cases). [5]
PIK3CA mutations have been found at a frequency of 2-7% in NSCLC (in exon 9 and exon 20). [6]
For the configuration of the Plasma-SeqSensei™ NSCLC RUO kits, four key cancer genes were selected after comparison of mutation frequencies using the COSMIC (Catalogue of Somatic Mutation in Cancer) and cBioPortal for cancer genomics databases.
Feature
- Highly sensitive <0.07 % MAF
- Able to detect 7 MM with 95 % confidence across all mutations (absolute quantification)
- High flexibility of between 2 - 16 samples/run
- Fast TAT (2 days)
- Convenient software
References
Chae et al. Detection of Minimal Residual Disease Using ctDNA in Lung Cancer: Current Evidence and Future Directions. Journal of Thoracic Oncology 14(1), 16-24, 2018.
Schwartzberg et al. Liquid biopsy mutation panel for non-small cell lung cancer: analytical validation and clinical concordance. Precision Oncology 4(15), 1-7, 2020.
ESMO. EGFR IN LUNG CANCER: ESMO BIOMARKER FACTSHEET. 2015.
Lee et al. Precision treatment for metastatic non–small cell lung cancer: A conceptual overview. CLEVELAND CLINIC JOURNAL OF MEDICINE 88(2), 117-127, 2021.
Veluswamy et al. KRAS G12C-mutant non-small-cell lung cancer: biology, developmental therapeutics, and molecular testing. The Journal of Molecular Diagnostics, 2021.
Wang et al. Clinical Significance of PIK3CA Gene in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis. BioMed Research International, 1-9, 2020.
https://cancer.sanger.ac.uk/cosmic
https://www.cbioportal.org/