Y Chromosome Deletion Detection System, Version 2.0
Standardized Screening Panel Amplify key functional regions associated w/Azoospermia Factor (AZF)
For Researchers Studying Y Chromosome Deletions
The Y Chromosome Deletion Detection System, Version 2.0, provides a standardized screening panel amplifying only informative nonpolymorphic sequence tag sites (STS) on the human Y chromosome. The system amplifies key functional regions associated with Azoospermia Factor (AZF), namely the regions that flank AZFa and cover AZFb, AZFc, AZFd including DAZ, KAL-Y, SMCY and flanking loci for other key spermatogenesis-related genes (namely RBM1, DFFRY and DBY).
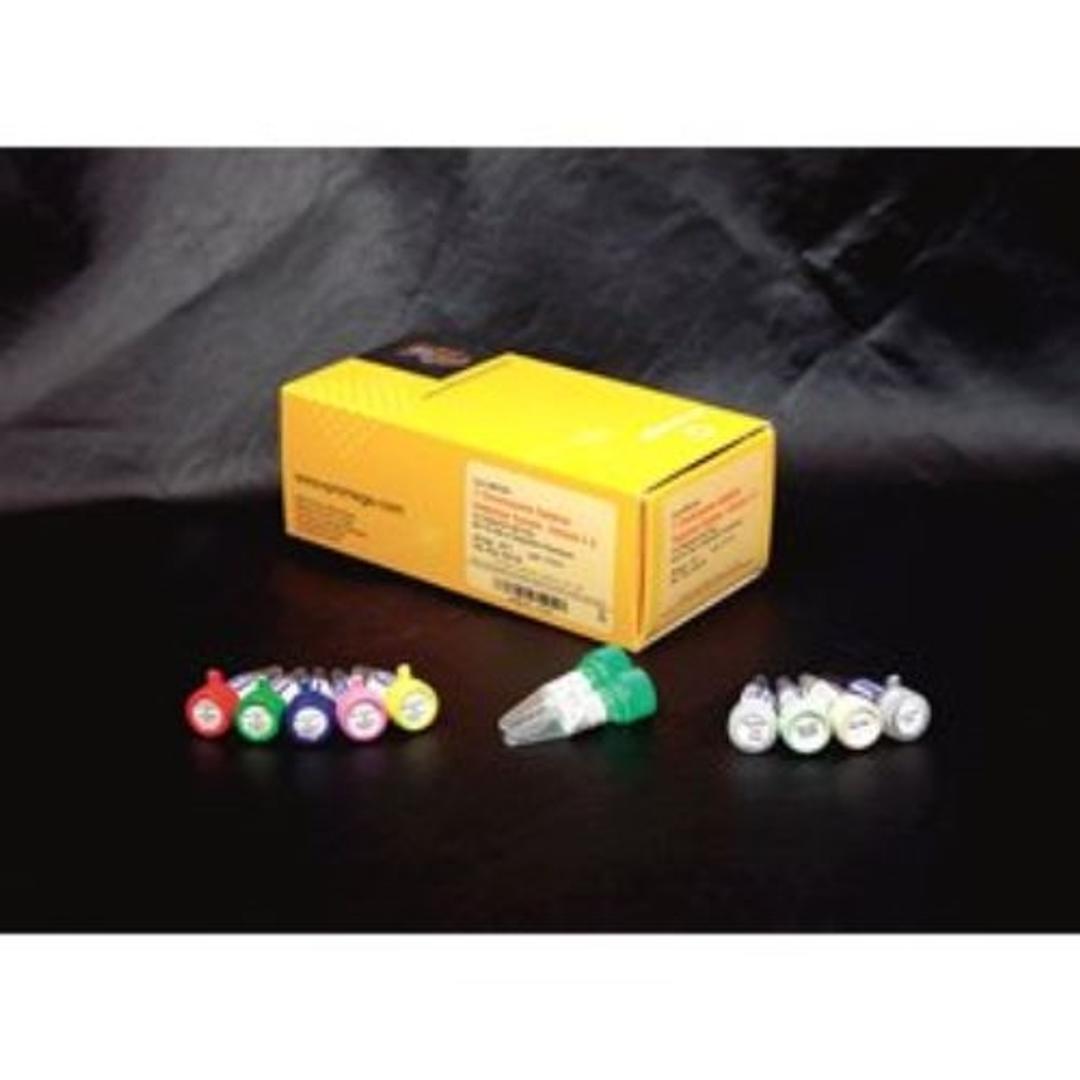
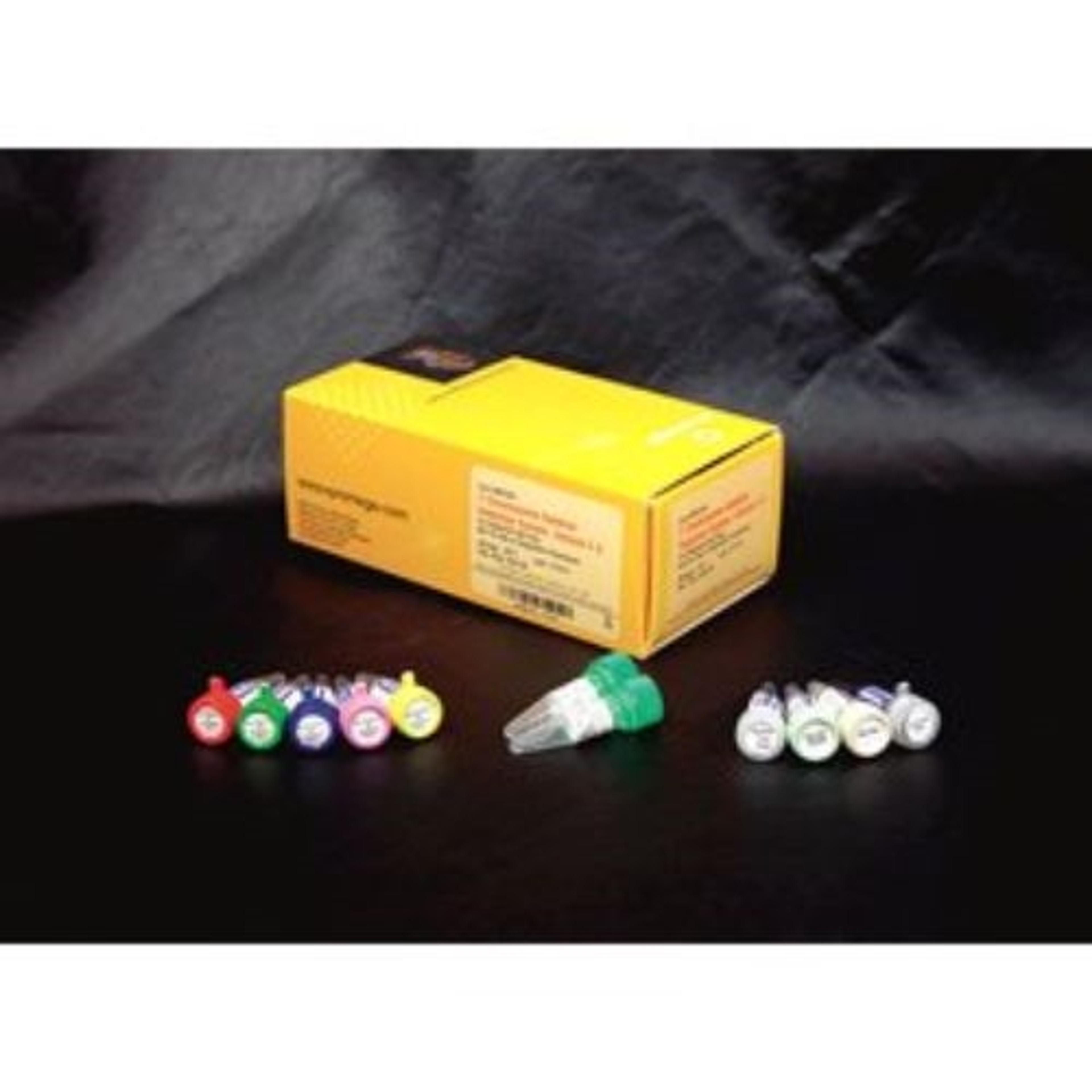
Five Multiplex Master Mixes, with a total of 20 characterized Y-specific primer pairs, are included. Four of the multiplex primer sets contain a control primer pair that amplifies a fragment of the X-linked SMCX locus. One of the multiplex primer sets (Multiplex E Master Mix) contains a control primer pair that amplifies a unique region in both male and female DNA (ZFX/ZFY). Finally, a primer pair that amplifies a region of the SRY gene has been included in Multiplex E Master Mix as a control for the testis-determining factor on the short arm of the Y chromosome to detect XX males arising from Y to X translocations.
The Multiplex Master Mixes are designed to facilitate the simultaneous amplification of several different regions of the Y chromosome. The amplification products (83–496bp) of the five multiplex PCR amplifications can be clearly separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
Failure to amplify specific regions of the Y chromosome is indicative of Y chromosome deletions in the test sample. The size control ladder provided minimizes analysis time and the possibility of misinterpreting molecular weight of amplification products.
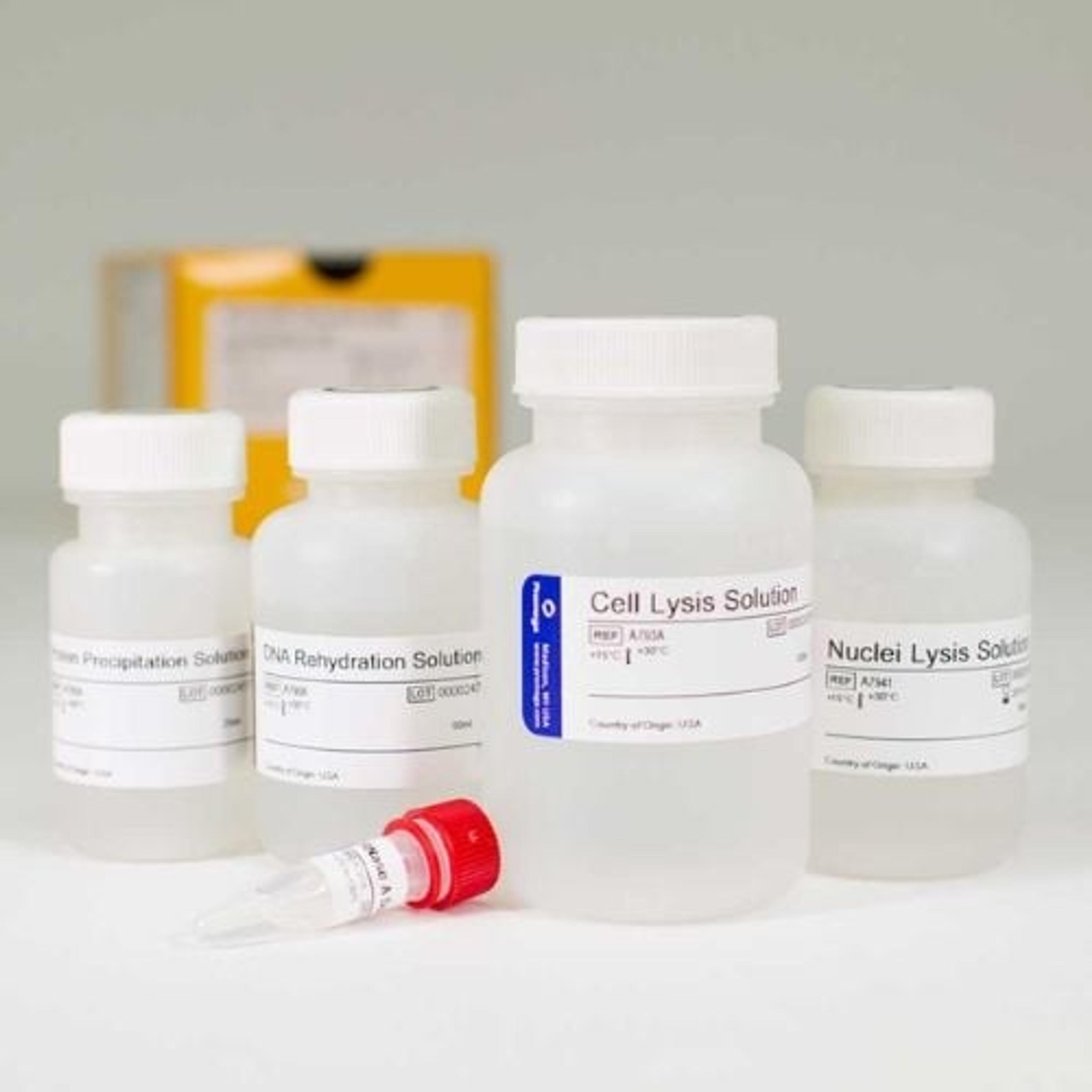
The Y Chromosome Deletion Detection System is easy to use and provides the flexibility to amplify genomic DNA purified using various methods and with a PE480 (oil overlay) or PE9600/9700 (non-oil overlay) thermal cycler. The improved formulation and use of GoTaq® DNA Polymerase minimizes dropouts.
References
- Skaletsky, H. et al. (2003) Nature 423, 825–37. * Especially note Figure 2, available online as data supplementary to the article.
- Vollrath, D. et al. (1992) Science 258, 52–9.
- Foote, S. et al. (1992) Science 258, 60–6.
- Kent-First, M.G. et al. (1996) Nat. Genet. 14, 128–9.