A whole HIVE in your hand: How a standalone device can change how we utilize single-cell sequencing
The HIVE system is changing how samples can be stored for single-cell sequencing, opening up its potential as a technique to many more research studies
24 Mar 2022

Many research projects could benefit from the analytical data that single-cell sequencing provides but are stymied by challenges with the workflows involved. These include differences between stored and fresh samples, and damage to delicate cells caused by storage techniques themselves.
Honeycomb Biotechnologies set out to develop a product that would overcome these barriers to make single-cell sequencing a viable technique for a broader range of research studies. What they created was a portable, handheld, single-use device that enables gentle capture, robust storage, and easy processing for the analysis of single-cell samples. It is called the HIVE. In this interview, we speak with Dr. Irene Whitney, Director of Applications and Collaborations at Honeycomb Biotechnologies, about the HIVE and how it changes the way that samples for single-cell sequencing are stored and processed.
Preserving sample integrity for more reliable sequencing
The first problem Honeycomb Biotechnologies wanted to overcome was one of sample storage and eliminating the variance between stored and fresh samples. As Whitney explains, “Any time you're doing single-cell work, the gold standard is to be able to work with fresh samples, fresh cells. And if your sample collection site is removed, either in place or even in time from when you're doing the rest of your experiment, you have to utilize different sample storage methods – say something like cryopreservation to store and or ship your samples – or work with frozen tissue and then dissociate and get nuclei. But both of those have their own problems and can affect the quality of your sample.”
Cryopreservation is a common storage method that can be irreproducible and cause poor cell viability and cell stress due to ice crystal formation during freezing, leading to poor quality data. Samples of delicate cell types such as sperm can be damaged by cryopreservation, instigating the need for remedial efforts to preserve sample integrity1.
The HIVE integrates sample storage into the workflow. After a sample is collected and loaded into the device, a cell preservation solution is added to stabilize the RNA until you’re ready to process. This novel sample preparation is now leveling the playing field for fresh versus shipped samples. Whitney adds, “We generated data comparing fresh samples versus those stored on the HIVE, and we see that they're equivalent in terms of the quality. So, it gives you access as if you had been working with fresh tissue, but you've overcome some of these collection issues, which may have been a barrier before.”
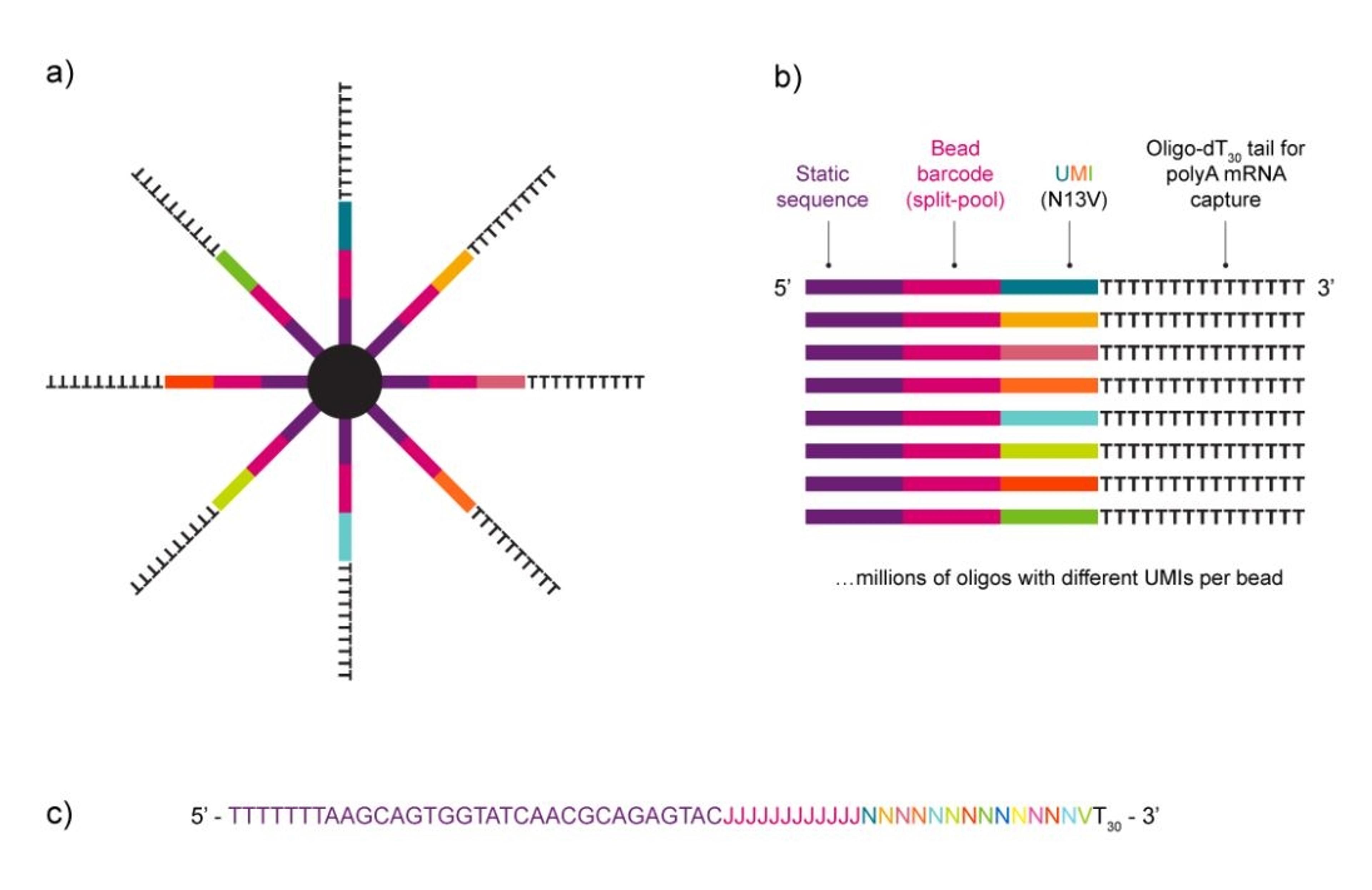
As a portable, hand-held device, the HIVE is also easy to use. You load your sample into it and single cells filter into pico wells following a Poisson distribution. When the cells are separated, you add in the cell preservation solution and then either store your sample or continue the workflow to process it.
Once in the pico wells, the cells settle with mRNA transcript-capture beads that are already present in the well. When you’re ready to process your cells, the pico wells are sealed, forming chambers with one cell and one bead. Lysing the cells allows mRNA to bind to the beads, which are then easily collected for sequencing. As Whitney says, “after you've captured your transcripts, we just spin the HIVEs to get a bead pellet with all of your captured transcripts and then move to a 96-well plate format for the rest of library prep.”

Fostering feasibility with single-cell sequencing
The HIVE aims to overcome barriers faced in sample preparation and storage to open up single-cell sequencing as an option to new research areas and studies. For some of these studies, single-cell processing wouldn’t have been possible due to difficulties with variable storage time, variable storage protocols or sample collection at different sites. Whitney explains, “I think we see the HIVE as enabling a much broader range of researchers to use single-cell in their research. It’s sort of democratizing single-cell sequencing.” She continues, “The other side of the technology that enables that is the fact that it works independently of any specialized equipment. The HIVE is just a single-use handheld device that's shippable, and the entire workflow can be done with standard lab equipment that you'd find in any molecular biology lab.”
We see the HIVE as enabling a much broader range of researchers to use single-cell in their research.
Dr. Irene Whitney Honeycomb Biotechnologies
Time-course studies, such as those used in drug discovery, are just one example that may now see single-cell sequencing as an option. These types of studies require collecting and running discrete experiments on large sample numbers at various time-points, which not only are labour-intensive, but also increase the risk of variability each time the samples are managed. Because the HIVE’s unique storage capability allows recovery of complete biological information, including from fragile cells, researchers are granted newfound flexibility for sample handling without introducing another variable that could compromise data quality. As Whitney attests, “With the HIVE you are able to batch samples together once you've collected all of them, with consistent results between those stored for different amounts of time.”
In the future, Honeycomb Biotechnologies will be looking at other aspects of single-cell analysis workflows to see where they may be able to support research efficiencies. Whitney elaborates, “the HIVE is sort of at the center of a workflow. But it's important to remember that there are things upstream and downstream of the HIVE that are important to have in place and well-established so that someone can complete their entire workflow.” Upstream of the HIVE, single-cell preparation remains the major source for technical variation and batch effects, where cell dissociation and any enrichment required by rare cell types pose their own unique specifications to yield meaningful results. Downstream of the HIVE, analyzing the data can be difficult to interpret if the study wasn’t designed appropriately2. The HIVE bridges the two sides of the workflow by optimizing sample storage. Beads manufactured by LGC, Biosearch Technologies enable the quantitative readout of transcripts that are analyzed by Honeycomb’s custom software solution, BeeNet™. As Whitney explains, “Honeycomb has been working closely with LGC. Suppliers that focus on supply chain security are important partners for our HIVE components.”
Ultimately, Whitney and the team at Honeycomb Biotechnologies want to continue to help people to access single-cell sequencing for their studies where previously it may not have been viable. Whitney concludes, “Single-cell analysis is not a cheap endeavor. All of the innovations of the HIVE and our future areas of focus will lead to making the most cost-effective experiment possible, where you have the best viability, you have the right number of cells, and you've enriched for the population that's most important to you. We want to make it so you're not spending those sequencing dollars on poor quality cells, or cells that are not biologically important to the questions that you're asking.”
References
Zhang R et al., (2022). Resveratrol and lycium barbarum polysaccharide improve Qinling giant panda (Ailuropoda melanoleuca Qinlingensis) sperm quality during cryopreservation. BMC Vet Res 2022 Jan 7;18(1):23. doi: 10.1186/s12917-021-03122-2
Nguyen Q et al., (2018). Experimental Considerations for Single-Cell RNA Sequencing Approaches. Front. Cell Dev. Biol., 04 Sep 2018. doi: 10.3389/fcell.2018.00108