NxSeq™ Single-cell RNA-seq Beads
Consistent, high-quality beads for single-cell mRNA sequencing applications using Drop-Seq methods
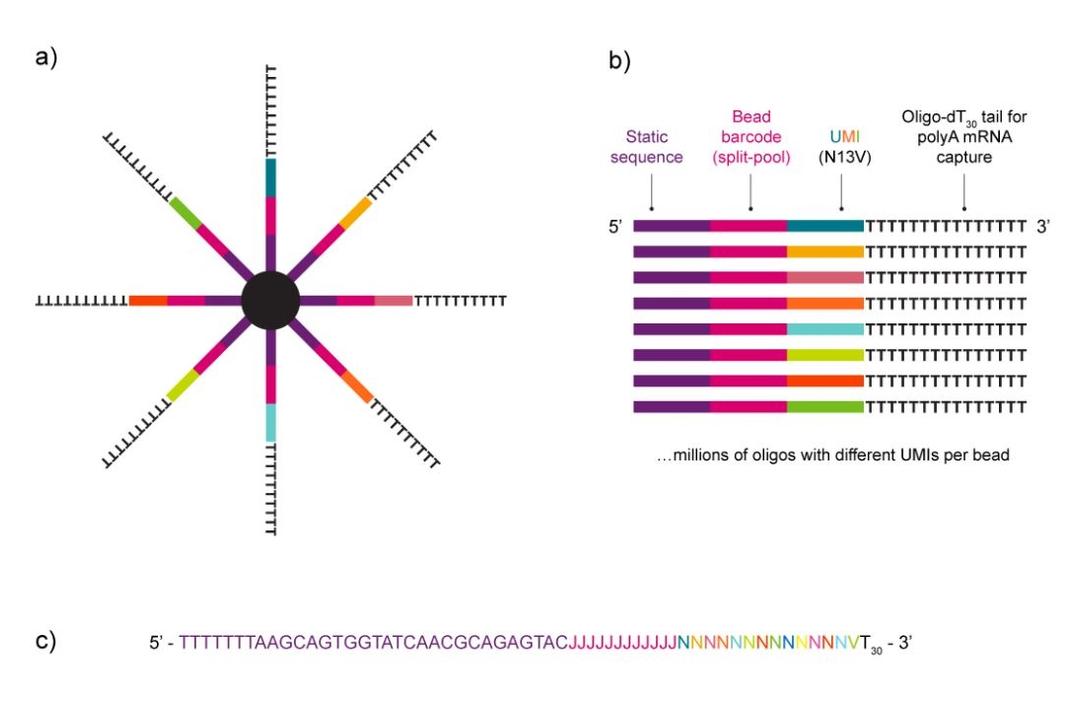
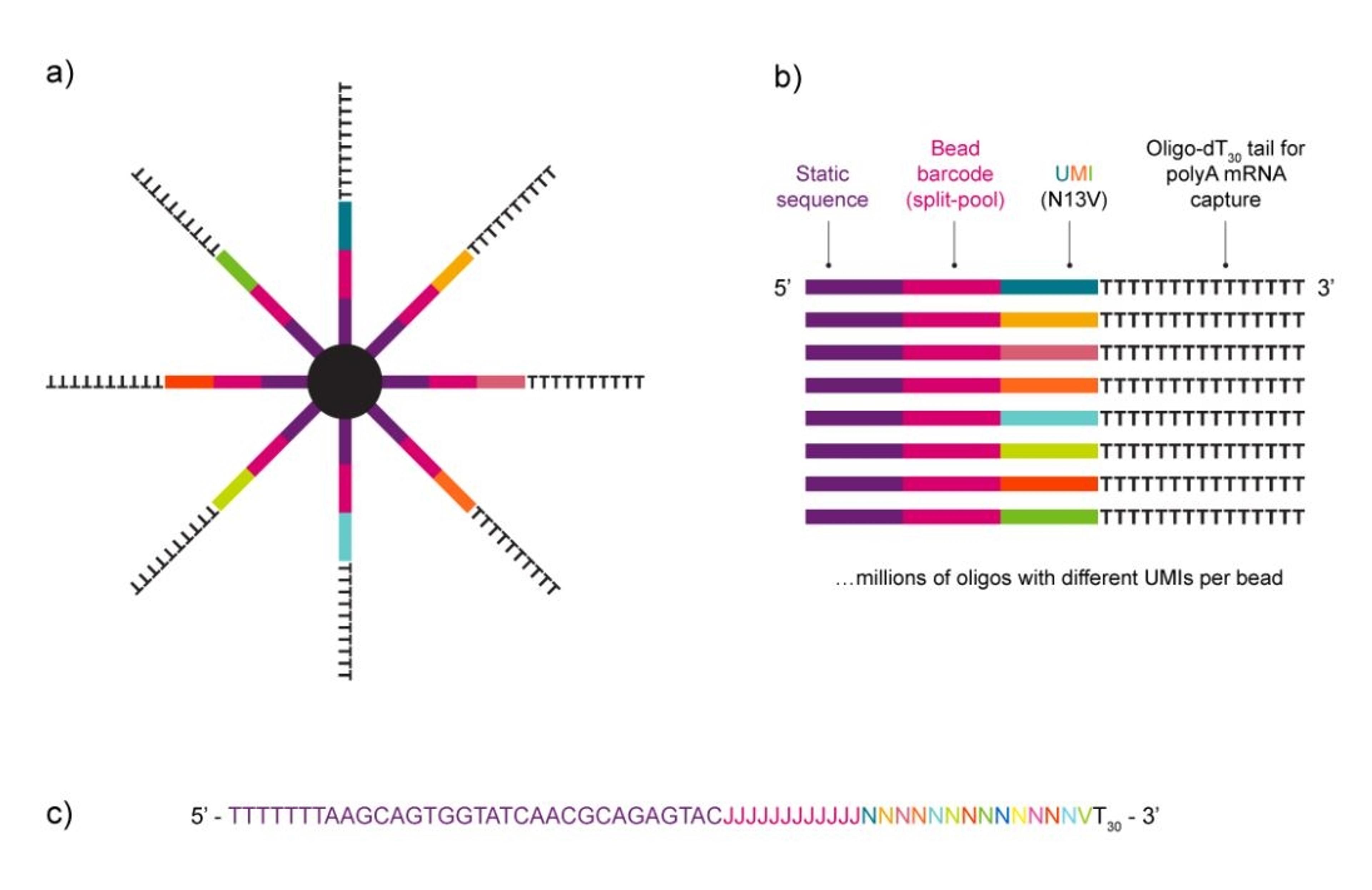
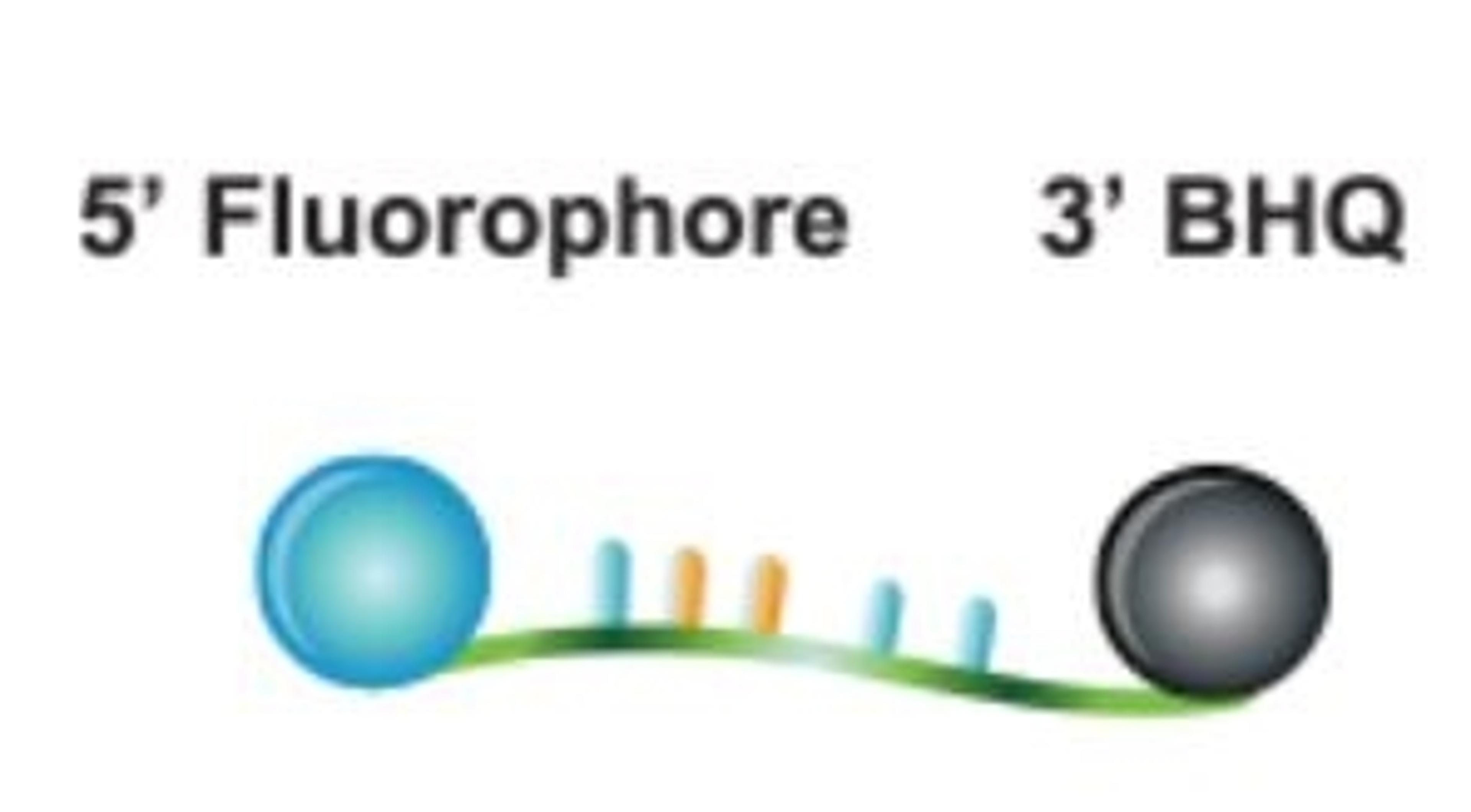
NxSeq™ Single-cell RNA-seq beads contain a covalently attached library of barcoded oligo-dT capture oligonucleotides to enable single-cell mRNA sequencing experiments using Drop-seq1,2 methods or similar instruments. Every bead-attached oligo contains a 12 base, bead-specific barcode, a 14 base unique molecular identifier (UMI) that is distinct between oligos, and a 3’ 30 base, oligo-dT mRNA capture sequence.
- Consistent: Each lot of beads has tightly controlled bead size and produces consistent performance lot to lot.
- Powerful: The 14 base UMI enables sequencing and identification of more unique reads.
- Quality-controlled: Each batch of beads is functionally tested to ensure you get the consistent performance you require from experiment to experiment.
- Flexible: Custom dispensing and pricing available for bulk, high volume purchases.
References
1. E.Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar, M. Goldman, I. Tirosh, A. R. Bialas, N. Kamitaki, E. M. Martersteck, J. J. Trombetta, D. A. Weitz, J. R. Sanes, A. K. Shalek, A. Regev, S. A. McCarroll, Cell, 2015, 161, 1202-1214.
2. http://mccarrolllab.org/dropseq/