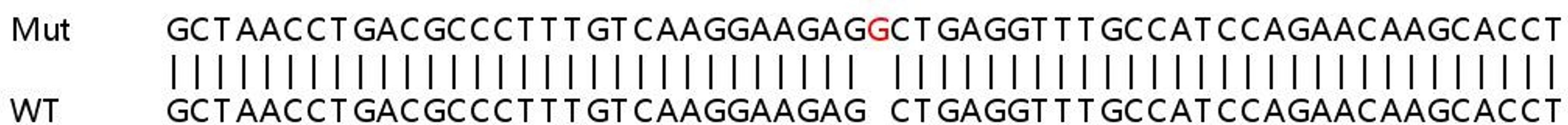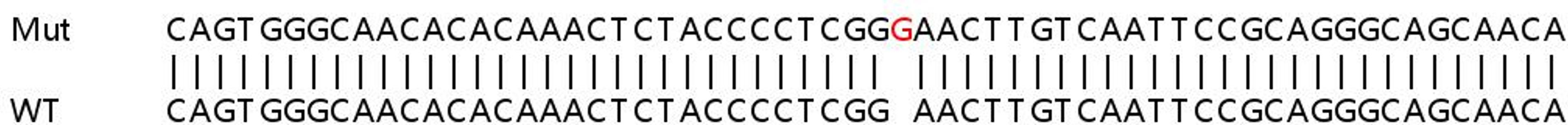Breakthroughs in guide RNA: Creating successful CRISPR-Cas9 knockouts
Abcam scientist Dr. Yongwon Kwon discusses a new method that could help scientists simplify their gene-editing processes and extend the validation of cancer cells
28 Jun 2020

As an exciting area of life sciences research with a vast and quickly evolving potential, CRISPR-Cas9 gene editing remains a relatively new technique that still faces a number of significant hurdles, including highly variable knockout efficiency, off-target cuts and safety concerns. Abcam scientist, Dr. Yongwon Kwon and his expert gene editing team, are focused on overcoming these challenges to produce high-quality research tools, with a personal focus on designing better guide RNAs (gRNAs) for increased gene-editing success.
In this exclusive SelectScience interview, Kwon shares insights into Abcam’s advanced CRISPR-Cas9 technology, and looks ahead to the future with a new, highly-efficient protocol for gene knockout (KO) in primary cancer cells. This novel, optimized approach makes it possible to edit multiple genes simultaneously — saving both time and resources while simplifying the gene-editing process for next-generation immunotherapies.
Tell us about your role at Abcam
YK: I am a scientist leading the molecular group in the cell engineering team at Abcam. Currently, my role is to design gRNA and donor DNA for CRISPR-Cas9 genome editing, including knockouts, conditional knockouts, base editing, conditional expression, point-mutation and knock-in (KI) techniques.
What inspired you to get into the world of science?
YK: When I was 11 years old, my grandmother passed away from cancer. This inspired me to be a scientist so that I could help advance cancer therapies. Over the past 10 years, I have been working to identify biomarkers in lung and breast cancers. In the future, I hope our work here can help researchers contribute to the development of gene therapies for the treatment of cancer.
What are the benefits of optimizing Cas9 RNP transfections?
YK: CRISPR-Cas9 techniques offer relatively simple molecular tools for genome editing. The CRISPR-Cas9 complex consists of a gRNA and Cas9 CRISPR-associated protein 9 nuclease, which together form a ribonucleoprotein called an RNP complex. Based on the biochemical ability of this complex, it can bind and cleave the target site.
In the past, many researchers chose a plasmid to deliver gRNA and Cas9 to their cells for editing, because plasmids are inexpensive, easy to work with, and they can include selectable markers. However, there are several problems associated with expressing gRNA and Cas9 as plasmids. The first of which is that the introduction of CRISPR components as plasmids is often associated with high levels of ‘off-target effects’ or unintended edits. Because plasmids can last up to several weeks inside cells gRNA and Cas9 is expressed continuously, this increases the chance of the gRNA-Cas9 cutting the cell’s genome in the wrong place.
When CRISPR-Cas9 are delivered in a pre-complex RNP format, the chances of off-target mutations are lower, as RNPs are only active inside cells for a limited time. The active RNP complex will repeatedly cleave DNA strands at the target site and will only persist in transfected cells for about a day before being degraded. As a result, fewer off-target cuts are made.
Secondly, plasmid DNA may randomly integrate into the host genome of target cells. By using the RNP format, it avoids the risk of DNA integration. Another benefit of using RNP is that, in this format, the gRNA is protected by Cas9, so it is less prone to degradation. I use synthetically made gRNAs because they can be chemically modified to protect them from endonuclease degradation and immunological attack. Both these factors enable the gRNA to target genomic DNA more efficiently during the short time they are active within the cell.
What current challenges are scientists facing with this technique?
YK: The safety and efficiency of this technique are important concerns that require comprehensive study. The efficiency of a CRISPR-Cas9 system can be impaired by off-target alterations. Specifically, gRNA tends to have a relatively high mismatch tolerance and Cas9 often cleaves off-target sites that have sequences similar to those of the target genes. To increase the specificity of Cas9 and improve gRNA design, researchers have generated Cas9 “nickase”. Nickases are highly accurate and can create single-strand rather than double-strand breaks. Furthermore, when used with two adjacent gRNAs, they can lower the probability of off-target editing.
There are several gRNA scoring prediction algorithms available for researchers to use today. Many of these tools calculate on-target and off-target scores, based on the algorithm developed by Doench et al. However, we still need to improve this algorithm because the current on-target and off-target predictions are different from the actual cutting efficiency.
Another improvement in the potential application of CRISPR-Cas9 technology is with the use of novel Cas9 enzymes. A protospacer adjacent motif (PAM) sequence is necessary at the target site for accurate CRISPR-Cas9 genome editing. The existing Cas9 only recognizes a few of the many potential PAM sequences, limiting the application of this technology. Recently, a newer Cas9 has been developed that recognizes a broader range of PAM sequences, expanding the application of the technology.
Finally, homology-directed repair (HDR)-mediated KIs show a very low efficiency, just below 5%. Significant effort has been devoted by CRISPR researchers to increase the KI efficiency by improving target strategies, particularly for the insertion of large DNA fragments. This is also something that we are currently working on in our labs.
This led to an 85% increase in KO efficiency, compared with targeting with a single gRNA.
Dr. Yongwon Kwon Abcam
Tell us about your multi-guide RNA method and its benefits
YK: To improve the efficiency of CRISPR-Cas9 at KO, we have developed an optimized Cas9 RNP transfection method in which multiple gRNA are transfected together. A single gRNA at the target site may or may not result in loss of protein expression. However, combined transfection of multiple gRNAs per target would likely increase the chances of knocking out protein expression, as gene editing in this way allows very precise fragment deletion.
We have optimized our protocol for Cas9 RNP transfection using two guides per target. This led to an 85% increase in KO efficiency, compared with targeting with a single gRNA. An additional benefit of this technique is that it alleviates the need for such extensive gRNA selection, which saves valuable time.
We have applied this method to cancer cell populations and discovered that Cas9 RNP transfection eliminates the need for cloning and viral transduction, making functional studies of genes involved in cancer development and progression much simpler.
What do you see as the future developments in this area?
YK: CRISPR-Cas9 technology is now routinely applied for efficient gene editing, contributing to advances in biomedical research. Recently, CRISPR-Cas9 has been utilized for the identification of potential therapeutic targets and has even entered into clinical testing. It is important that safety and efficiency are closely monitored during these clinical studies, as this technology has the power to shape future clinical applications.
Hear more from the knockout experts:
• How to achieve research success with knockout models – Dr. Hanna Dreja
• Speeding the transition from bench to bedside with CRISPR/Cas9 – Dr. Karine Enesa
Want to know how to effectively harness KO validation for reproducible research? Register for this free webinar from Dr. Hanna Dreja>>
References:
Doench, J.G., et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene. Nature. 2014
Doench, J.G., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature. 2016