Call to action: Ensuring safe cannabis and cannabinoid products
Visiting Professor at the Johns Hopkins University School of Medicine demonstrates comprehensive pesticide testing methodologies to ensure cannabis product safety
18 Aug 2021
Hundreds of manufactured pesticides lead to very complex testing needs across the food and agricultural industries. However, only a small subset of the available pesticides has been regulated in the cannabis industry, and the target lists and action (not to exceed) levels vary from state to state and country to country.
Another caveat to pesticide testing is that only the specific compounds defined in the tandem mass spectrometry method can be identified and quantified. Any pesticides not defined in the instrument method are excluded from the analysis. This reality opens the door for the use of non-regulated pesticides since their presence in a product is unlikely to be determined.
In this on-demand SelectScience webinar, Dr. Anthony Macherone, Senior Scientist at Agilent Technologies, and a Visiting Professor at the Johns Hopkins University School of Medicine demonstrate analytical methodologies for comprehensive targeted and semi-targeted pesticide analysis to better ensure the safety of cannabis products, where legalized. Including, how comprehensive pesticide testing can be used to harmonize regulatory needs across regions.
Register now to watch the webinar at a time that suits you or read on to find highlights from the live Q&A session.
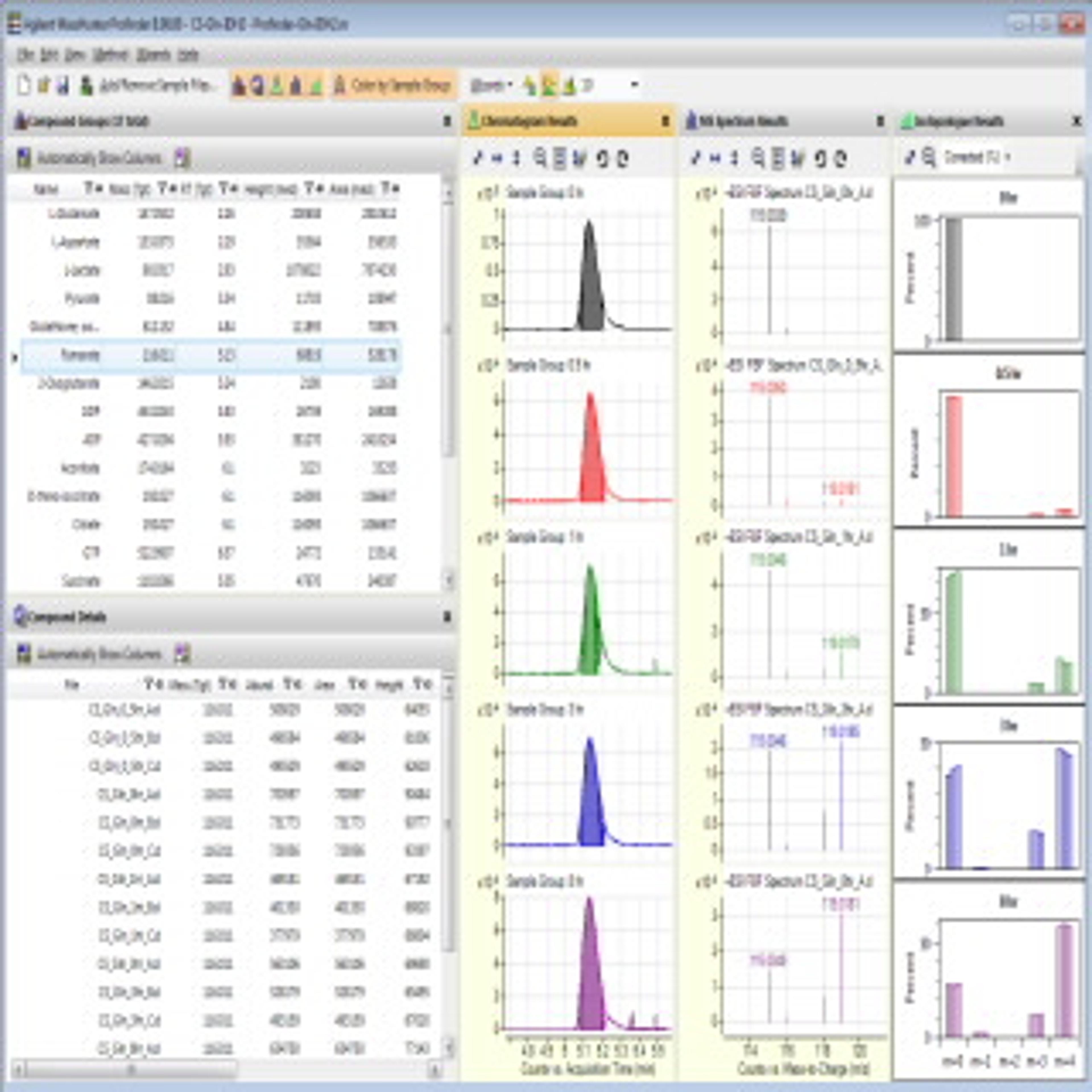
Watch on demandQ: What is the difference in the data when collected with tandem quadrupole mass spec or time-of-flight mass spec?
AM: Tandem quadrupole, or MS/MS, methodologies are targeted methodologies where the target list is defined within the method. But, if there is a pesticide in that sample that is not on your target list, you won't see it because it's not defined to be acquired, to be identified, to be quantified - that's how MS/MS works. Time-of-flight information, essentially, collects all chemical features that are in that sample.
If there is a pesticide at a level that can be identified with the high-resolution accurate mass system, then you will see it, and it may be a pesticide that is not on a specific state or region list. Therefore, this gives you more comprehensive information about that sample. You can target and quantify the known list on a time-of-flight instrument, but you can also seek and interrogate the data for other compounds, as I showed in the presentation.
Q: Normally, benomyl is converted into carbendazim. Is there any way that we can detect both separately in the same static sample?
AM: Absolutely. These are two compounds, benomyl is the larger of the two in terms of the empirical formula. Essentially, it loses a butyl amide moiety, I believe, resulting in carbendazim. You can certainly differentiate those compounds from one another using time-of-flight information. Potentially with MS/MS as well because they are very different chemicals.
There is this loss of a pretty large piece of the molecule as you go from benomyl to the other compound. If this conversion occurs while you prepare your sample and it is sitting on the instrument waiting to be analyzed (so pre-chromatography), you most likely would see them at two different retention times. You could again target them with MS/MS, based on very specific precursor product ion pairs. At time-of-flight, certainly, you would be able to differentiate them by retention time.
But if this occurs in source (everything post-chromatography), these two compounds will be right on top of each other. They will essentially coelute, and so the carbendazim will be right underneath the benomyl. However, you still could differentiate them with time-of-flight because they have a very different empirical formula, and so you could use the high-resolution accurate mass to distinguish those, even though they are right on top of each other. MS/MS could do this as well.
Q: Can time-of-flight mass spec be used for quantitative analysis?
AM: I think that is what I wanted to show with this comprehensive screening, something called "semi-targeted analysis". For example, when you are using your LC/time-of-flight instrument and collecting all the information from a particular sample, you can target all those cannabinoids, 11 or 17 for example, and you can create a pre-prepared calibration curve that you can quantify against.
Therefore, you certainly can do quantitative analysis using time-of-flight mass spectrometry, and you can do this in a semi-targeted way. Because, again, when quantifying those cannabinoids, you're also going to see all these other great things, like flavonoids, which you could quantify too if you wanted to.
Q: How are unknown chemicals in time-of-flight mass spec identified?
AM: With time-of-flight information, you have your raw data, you have all of these potential chemical features in your chromatogram, little peaks here and there, sometimes even things you don't see that are down in the baseline, but greater than the noise. You have to use very specialized software tools to extract those chemical features appropriately so you can distinguish them from the noise, identify what's real, discard what is not real.
There are commercially available tools. At Agilent, we have the LC/time-of-flight information software called MassHunter Profinder Software. Profinder goes through and performs molecular feature extraction in a number of different ways, pulling those real chemical features out of those chromatograms and giving them in a format where you could possibly search those against known spectral libraries, like Methylene or NIST. There are online tools such as XCMS that also do this chemical feature extraction.
On the GC side, at Agilent, we use another software platform, MassHunter Unknowns Analysis, which does chromatographic deconvolution. This is a very well-known way, going back to AMDIS, which was developed by the government to identify chemical warfare agents, so they are very sophisticated. Again, it extracts the real features from the raw data using chemically chromatographic deconvolution. But, you need to have some software tools to really go in there and do this for you.
Certainly, you could search out a specific ion, given plus or minus 10 parts per billion, million or similar, and extract those out looking for things by hand, but that's certainly not a very efficient way to do that. You really do need to use these software tools, although they are somewhat different – for example, gas phase using electron ionization versus liquid phase using electrospray.