‘Google Maps’ for the human body – How spatial genomics is revolutionizing our understanding of disease
Discover how the Wellcome Sanger Institute is helping to develop a physical atlas of all human organs
18 Feb 2024
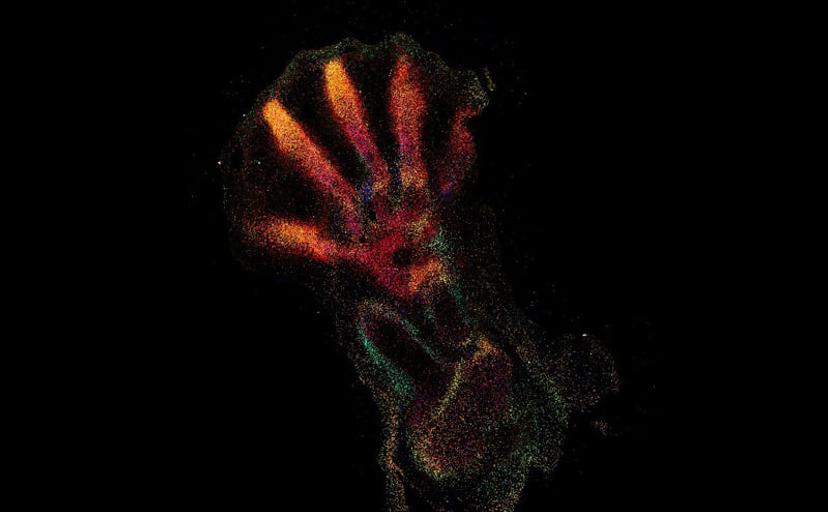
The Human Cell Atlas is an international consortium with the shared ambition of discovering and documenting the rich diversity of cell types across different human tissues. Understanding cell-type diversity throughout the human body allows researchers to explore how these cell types function and dysfunction in human health and disease states.

“A lot of the disease symptoms that span many different organs arise from the same specific cell types in the human body,” explains Dr. Omer Bayraktar, junior group leader at the Wellcome Sanger Institute, a leading player in the consortium. Bayraktar highlights that neurological disorders illustrate this phenomenon incredibly well, due to the rich diversity of both neuronal and non-neuronal cell types in the brain. Examples of this include the dopaminergic neurons that are affected in Parkinson's disease and the interneurons in schizophrenia.
In this exclusive SelectScience® interview, we find out more about the Wellcome Sanger Institute’s contribution to the Human Cell Atlas, including the development of cutting-edge technologies to further understanding of the human brain and the role of cell diversity in neurological disease.
From genes to organs: Creating a 3D map of the entire human body
The first wave of the project makes extensive use of single-cell RNA sequencing (scRNA-seq) technologies, enabling researchers to see the molecular diversity and gene expression patterns within previously unstudied human tissues in a high-throughput fashion. However, through use of this dissociative transcriptomic method, the spatial context of cells within the tissue of interest is lost. Of the method, Bayraktar states: “This is a good tool for achieving an unbiased first look at the different cell types across different organs, but you have no idea where the different cell types are coming from in your tissues of interest.”
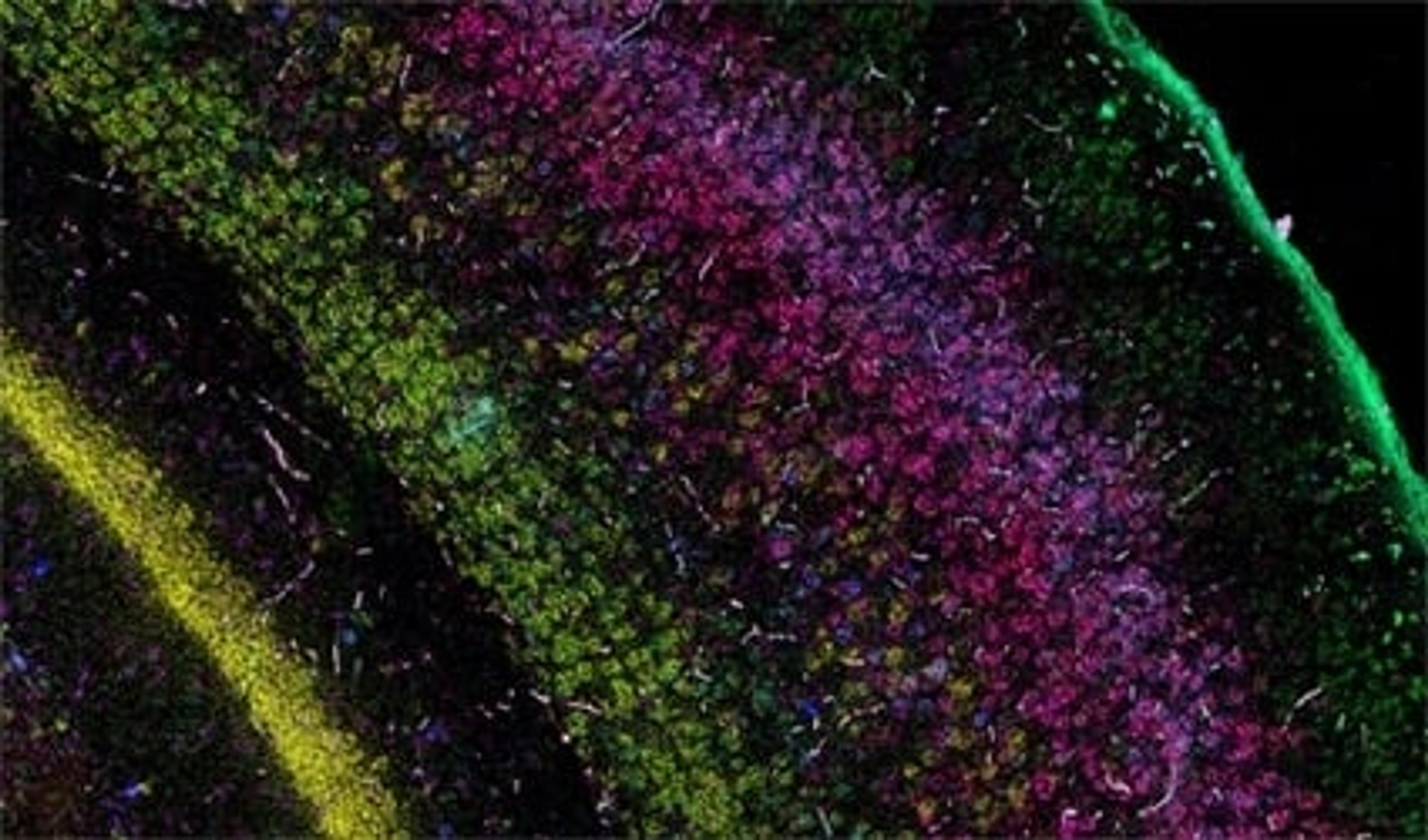
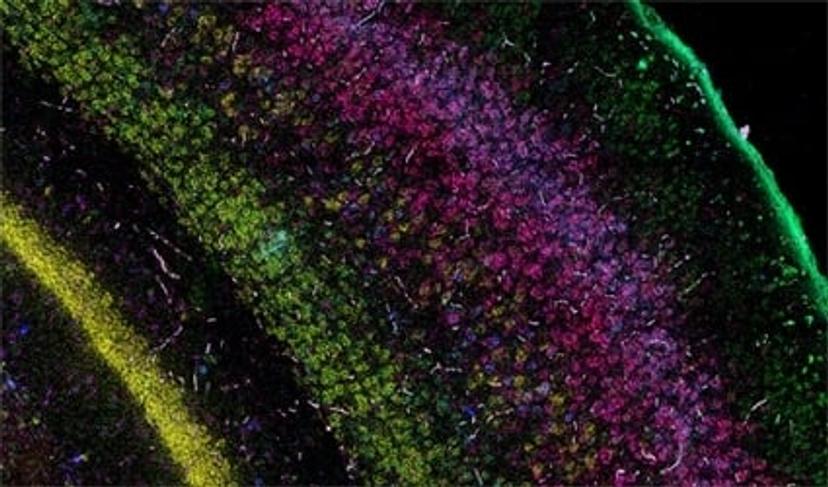
The current focus now takes this first-wave information and locates the identified cell types and expressed genes within the intact architecture of a specific organ. Bayraktar shares that the aim is to create, as many Human Cell Atlas labs like to say, “the Google Maps of the human body”. The project combines the strengths of anatomy and cell morphology with molecular profiling, to create a three-dimensional atlas to help identify where different cell types are located in the tissue, as well as what genes and proteins are being expressed.
This spatial interrogation of gene expression is known as spatial genomics, and despite the promise of this emerging field, the current methods available are still in their infancy.
Sensitive, robust, and versatile spatial genomic methods
When looking for imaging solutions for spatial genomics, Bayraktar’s lab favors commercially accessible technology that offers robust levels of detection and readily works on very many different tissue types: “We want more convenient ways to study our different tissues, instead of having to invest heavily on developing new technologies or troubleshooting these on each new tissue of interest.”
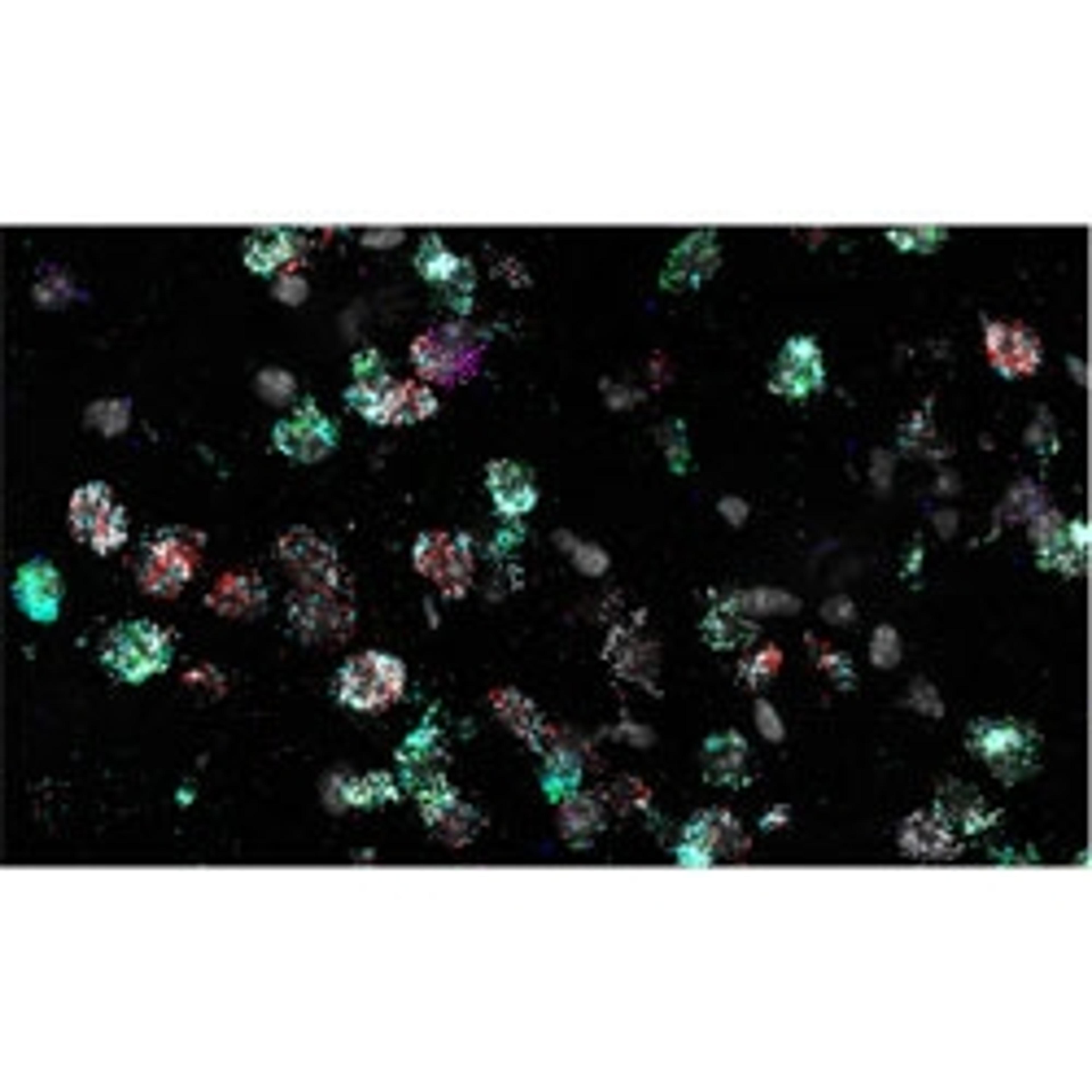
RNA in situ hybridization (RNA ISH) is one such imaging method that allows for the detection of the expression of chosen target genes of interest. Bayraktar and his lab use single-molecule fluorescence ISH (smFISH) and other RNA ISH techniques to perform highly sensitive, spatially resolved mapping of gene expression with single-cell resolution to validate the observations that come from their scRNA-seq studies.
“In our first study, we used low multiplex RNA ISH to map the previously unknown diversity of astroglial cell types across layers of the mouth and the human cerebral cortex,” says Bayraktar. “What RNA ISH allows us to do, at single-cell resolution, with very high transcriptomic sensitivity, is quantify the expression of our target genes of interest with spatial resolution.”
Scalability for the future study of human disease
Bayraktar highlights that the first wave of the Human Cell Atlas was very successful due to the highly scalable nature of scRNA-seq and transcriptomic methods. Due to the degree of cell-type diversity in human organs, there is still the need for this type of scalable platform for spatial genomic applications. “To chart the Human Cell Atlas of the entire body, we need access to large-scale platforms that can apply spatial genomic methods to human organs, of a large number of individuals, and ultimately the entire human body,” explains Bayraktar.
To be able to tell apart the different cell types across different human tissues, you need to be able to assay the expression of hundreds of genes simultaneously. The most robust RNA ISH platforms that are currently available address much lower levels of multiplex RNA detection. Bayraktar highlights the Wellcome Sanger Institute’s high-throughput spatial genomic initiative, aiming to “push the current level of multiplexing, taking it from imaging four genes to imaging several hundred genes simultaneously, and create scalable platforms that can be applied to many different tissue sections, organs, and ultimately the entire human body.”
Once these scalable spatial genomic platforms have been developed, they can be harnessed to identify the diversity and location of cell types throughout the body and relate this to gene expression patterns in disease states. The study of selective long-term cell durability, in disorders such as Down’s Syndrome or amyotrophic lateral sclerosis (ALS), is just one of the applications that Bayraktar and his lab hope to address. However, the ultimate aim is to apply this kind of technology to inform our understanding of a wide range of neurological, neurodevelopmental, and neurodegenerative disorders.
Looking for an RNA ISH solution for your gene expression studies?
Advanced Cell Diagnostics’s Pharma Assay Services provides RNAscope and BaseScope in situ hybridization (ISH) assay services to support pre-clinical and clinical studies for pharma and biotech partners globally. Tissue sectioning, ISH staining, high-resolution full slide scanning, scoring, and image analysis are performed by a dedicated team of highly trained specialists, scientists, and board-certified pathologists. With direct access to the developers of the technology, the Pharma Assay Services team provides unparalleled expertise in ACD’s ISH platform and delivers fast, high-quality data designed to meet your study objectives and timelines.