How well are your HCP ELISAs covered?
Discover the strengths and weaknesses of one of the most common types of coverage assay
13 Sept 2022
Validating host cell protein ELISAs is an essential part of biologics development. But what makes these coverage assays so vital, and what could we improve? Read on and see how Cytiva’s 2D DIBE approach is designed to combine the best of existing methods.
What is a coverage assay?
Coverage assays provide a means to validate the enzyme-linked immunosorbent assays (ELISAs) used in host cell protein (HCP) quantitation. They enable you to estimate the percentage of HCPs that can be detected, or ‘covered’, by ELISA, and direct you towards filling any gap. The goal is to develop processes for reducing HCPs, improving drug efficacy and patient safety, and meeting regulatory guidelines. These ELISAs and coverage assays are an essential part of process development in biologics manufacture, and can have an impact on the potential success of a drug.
Here, discover the strengths and weaknesses of one of the most common types of coverage assay, and learn about an approach that retains its best traits while enhancing speed and accuracy.
The need to stay covered in HCP quantitation
Bio-pharmaceutical companies have a duty to make sure their products meet safety regulations. Optimizing purification steps, and monitoring the presence and quantity of HCPs throughout by ELISA are two key ways to do this.
ELISAs are the workhorse of HCP quantitation, but they aren’t perfect. Raising antibodies against HCPs is not 100% effective, as not all HCPs are immunogenic enough to trigger a response in the animal host.
Given the potential for HCPs to affect patient safety, US and EU Pharmacopeia (USP 1132 and Ph. Eur. 2.06.34) state that ELISAs should also be monitored. This involves characterizing and validating the antibodies by coverage assays to check whether they have the expected sensitivity, specificity, and coverage.
Several biologics in recent years have shown immunoreactions in late clinical trials. These include Sandoz-Novartis Omnitrope™, IPSEN™ Coagulation Factor IX, and Genentech™ Lebrikizumab. This was a serious safety concern, and led to delays in their approvals.
As a result, 2016 and 2017 saw the US and EU pharmacopeia updated to include coverage assays as best practice for risk management for any future safety issues. Although these assays add some extra cost to product development, this is outweighed by the potential risk of delays in product approval.
The new guidelines recommend generic assay revalidation for every new batch of reagent, and process- and platform-specific assay revalidation after any change that could affect HCP composition.
Standard coverage assays and their challenges
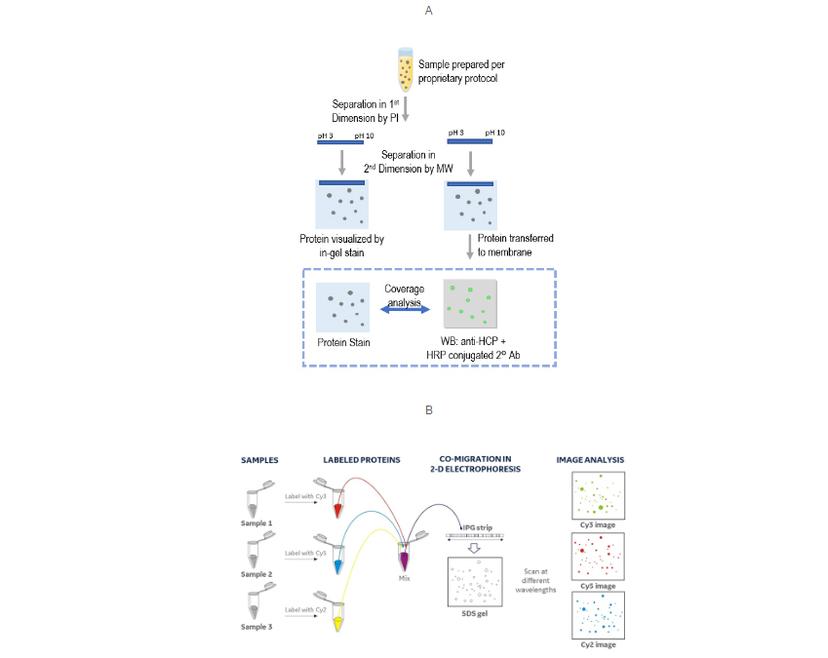
A coverage assay can be quite straightforward, often just 2D gel electrophoresis/Western blot. Comparing the colorimetric anti-HCP Western blot to a total protein stain, and identifying the matching spots, provides an estimate of antibody-to-HCP coverage.
This approach provides good resolution of individual HCPs as a 2D map, but has several challenges for something that’s so crucial to get right (USP 1132). These include:
- Denaturation of HCP antigens, which might not represent native antigen in ELISA
- Variability in electrophoresis and transfer efficiency between gels
- Reliability of matching spots between blot and gel
- Consistency between users in judging coverage
- Time required for experimental preparation and data analysis
So, what can we do about this?
The US pharmacopeia proposes using an immunoaffinity approach, followed by differential gel electrophoresis (DIGE) for checking coverage. DIGE is an established multiplexing variation of 2D gel electrophoresis/Western blot that enables you to separate and image up to three samples simultaneously, simplifying workflows and improving the reliability of spot analysis.
In this case, the method labels the column chromatography fractions: input (all HCPs), eluate (HCPs covered by anti-HCP antibodies), and flow-through (HCPs missed by antibodies), with different fluorophores. These fractions can run on a single 2D gel, benefiting from multiplexed fluorescence imaging and objective analysis by software.
If your lab currently uses 2D-gel electrophoresis/Western Blot for coverage assays, is there a simple way of gaining some or all the benefits of DIGE without changing approach?
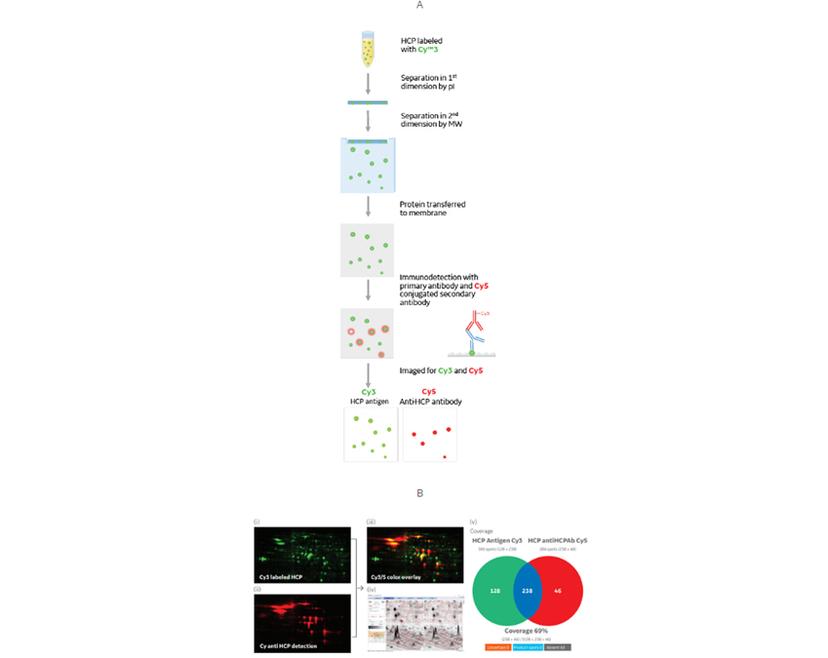
Improving speed and accuracy with 2D DIBE
If we take the immunodetection method used in Western blotting and combine it with the fluorescent tagging and imaging of DIGE, the result is 2D DIBE: differential in blot electrophoresis.
This assay format retains the high sensitivity of a primary/secondary antibody system, and the ability to multiplex using fluorescent dyes. When switching from a colorimetric Western blot, taking this approach allows you to run one gel rather than two.
Reducing the number of gels saves time and reagents and removes any variation from imperfect duplicate gels and transfers. Using a single sample of HCPs for detecting both total protein and anti-HCP immunoreacted spots also removes the risk of bias that could result from HCP loading differences between two gels.
2D DIBE/DIGE provides:
- Sensitivity: Sub-nanogram order of total HCP detection
- Accuracy: Resolution of a 2D map of HCPs
- Speed: Fewer gels needed compared to colorimetric Western blot
- Objectivity: Quantitative analysis of fluorescent signal by Melanie software
Making the switch to 2D DIBE
If you are currently using a standard 2D gel electrophoresis/Western blot coverage assay, this 2D DIBE workflow is no more time-consuming or challenging to set up. You just need to label your HCP samples with a fluorescent dye and swap the existing secondary antibody for a fluorescently-tagged antibody.
After that, you can benefit from the same speed of workflow, accurate software-driven quantitation, and cost-savings as the immunoaffinity/DIGE approach.
Written by Joe Hirano, Program Manager, Imaging, at Cytiva
