Demystifying the complex role of GABAergic neurons in sleep deprivation
Discover how cutting-edge single-cell RNA analysis technology is helping make strides in homeostatic sleep pressure research
1 Aug 2022

Single-cell RNA analysis is a powerful technology that enables the investigation of gene expression at the single-cell level. This provides the opportunity to explore, in depth, the activity and heterogeneity of cells and tissues. One scientist whose research has been revolutionized by this technology is Parris Washington-Chandler, a Ph.D. candidate in the Human Genetics program at Johns Hopkins University. Here, she works studying homeostatic sleep pressure in the lab of Seth Blackshaw as part of the neuroscience department.
In this exclusive SelectScience® article, Washington-Chandler shares insights into her work identifying and characterizing the GABAergic neurons involved in sleep deprivation. She ultimately hopes to reduce the public health burden of sleep deprivation disorders by uncovering the contributory mechanisms at a cellular level. Single-cell RNA analysis, using the RNAscope™ HiPlex manufactured by Advanced Cell Diagnostics (ACD), has played a key role in allowing Washington-Chandler to obtain and analyze large amounts of data, in a spatial and temporal context.
Parris Washington-Chandler recently presented her work in the SelectScience webinar series: Diversity, development, and dysfunction in the brain. Watch her presentation on demand, here >>
What burden do sleep deprivation disorders have on the general population?
PWC: It's been said that sleep disorders or chronic sleep loss is one of the most commonly overlooked health problems. An adult is supposed to receive on average 7 to 8 hours of sleep per night and I think we know just from our general experience that that often doesn't happen. The access to technology and the access to light at all times of day has also hindered our sleep health. That exposure to blue light, whether it be from your cell phone, your computer, etc. while you're trying to sleep has been affecting our bodies in the sense that they have affected our bodies' biological clocks and how we perceive light.
We often use the phrase biological clock in reference to women and their reproductive cycle, but almost every cell in your body has a molecular clock that's helping it keep time to know when to carry out a certain function. So, when we're exposing ourselves to all this light on a consistent basis, we throw off the timing of those clocks, so it carries out functions on an incorrect time scale. A large population of American individuals have issues with sleep, and it's not even documented. So, this is relevant to a larger public health issue, especially in this age of technology.
What is the overall goal for your research?
PWC: I study homeostatic sleep pressure. At its simplest, this could be described as your drive of tiredness, an increased tiredness as the day, or your time awake, proceeds. We're interested in identifying the neurons, specifically GABAergic neurons, that are responsible for sensing tiredness, and then further classify these as a cell type based on their pattern expression of molecular markers.
What impacts are you hoping to have with this work?
PWC: We hope that the impacts could be several-fold. One, we're interested in using the RNAscope™ HiPlex assay with ACD to map this area to get an idea of what these neurons are doing spatially. It would be great if we could regionally see that the clumps of neurons in this area are carrying out a specific activity and are expressing certain molecules in response to sleep deprivation. Now, it hasn't been that clear-cut, but overarchingly, we would like to identify and characterize those subtypes. Then, hopefully, use this information down the line for people who have issues with disorder of sleep.
The RNAscope HiPlex assay has been designed with the aim to expand researchers in situ hybridization potential. The technology allows simultaneous detection of up to 12 different RNA targets per slide-mounted sample. One major application for the RNAscope HiPlex assay is to validate gene signatures with spatial context at the single cell level. Here, Washington-Chandler describes how the RNAscope HiPlex assay has opened up limitless possibilities in her research.
What key challenges do you face when investigating these neurons?
PWC: One of the key challenges that we face is that this GABAergic population, particularly the GABAergic sleep-promoting population, is extremely heterogeneous and they're found throughout the brain, not simply localized in the hypothalamus. There are nitic oxide synthase sleep-promoting neurons in the cortex and sleep-promoting neurons in the ventrolateral preoptic area. There are also sleep-promoting GABAergic neurons in the lateral hypothalamus, which is the zone of the hypothalamus that I'm interested in. They are a large and diverse population and scattered throughout the brain. This makes them very difficult to study, especially if we want to categorize them by their secreted neuropeptide as there is a lot of overlap.
How do you use the RNAscope technology to help overcome these challenges?
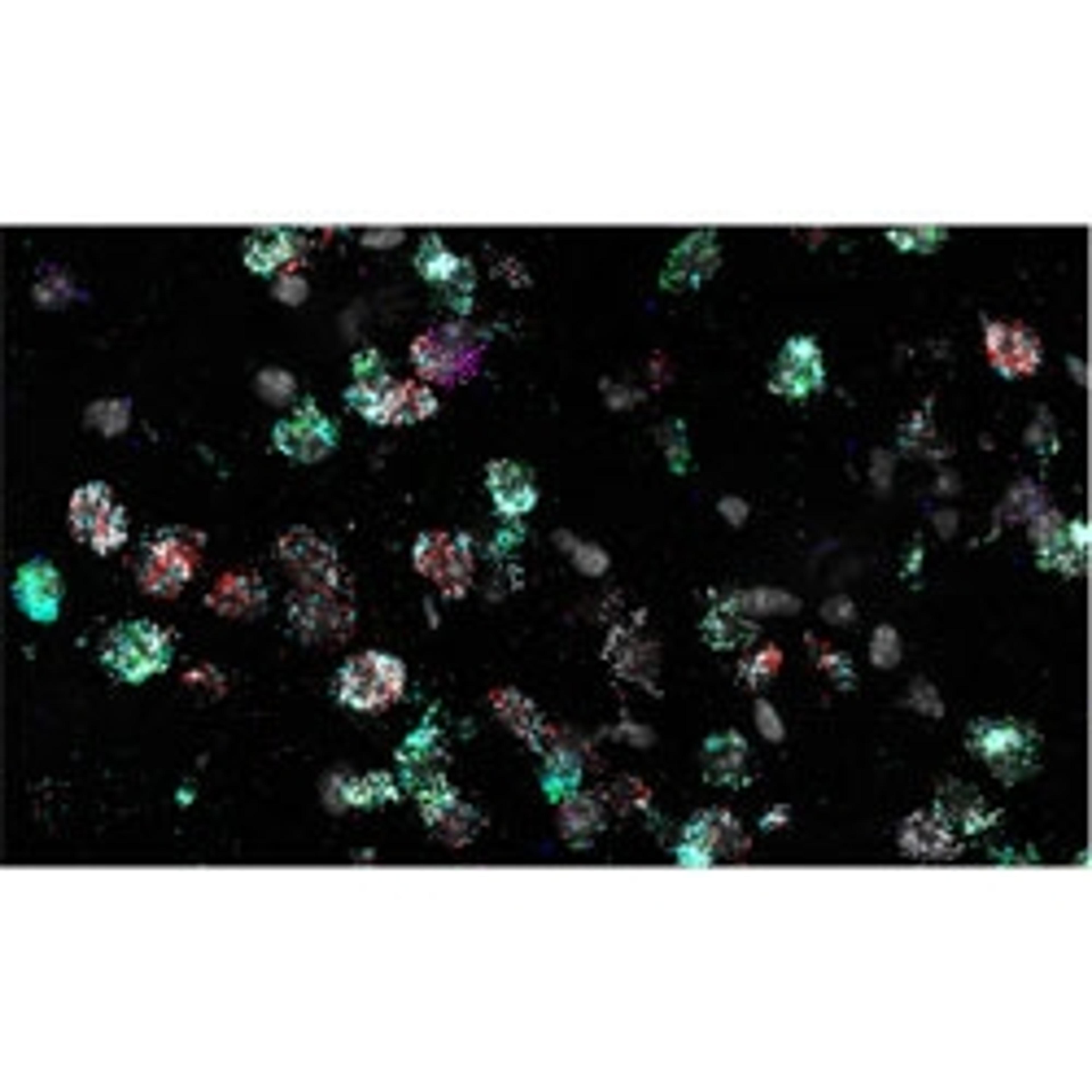
PWC: The RNAscope HiPlex is really cool for several reasons. It has both highlighted how diverse this population is, and simultaneously let us know that some things just might not be attainable. I have been working with 12 different probes to look at the state of the brain, and what probes are being expressed. I used a control population with no sleep deprivation, a sleep-deprived population where I've deprived the mice of sleep for 6 hours, and a group in which the mice are deprived of sleep for 6 hours, and then have a 3-hour period of recovery sleep.
One of our hypotheses for this was that we would be able to identify the specific spatial regions and these groups of neurons that are expressing this specific group of markers. HiPlex has been able to show me that the mechanism is not that clean and is very complex.
When we go to look at what these cells are doing in response to sleep deprivation, two neurons could be directly next to each other, but have extremely different expression profiles. It's not a clean grouping of cells responding in a certain way in one region of the brain and cells in a different region responding a different way. This reinforces that idea of heterogenous populations. We're seeing a lot of amazing things, but the great thing about HiPlex is that I can see what each individual cell is doing at that exact moment, for up to 12 probes. I can look at anything from almost any perspective dealing with these probes. Getting that level of detail and that amount of data is amazing.
It has been really useful to use this technology in combination with an automatic cell pipeline, called CellProfiler, to analyze the data. If you're interested in big data and collecting big data from single molecule fluorescence in situ hybridization (smFISH), this is a really good way to go.
What are you most excited about for the future of your work?
PWC: As a computational scientist and a data scientist, the sheer amount of information I've been able to get from using this technology is extremely exciting. I've been able to classify across 12 probes and combining that level of information is massive. For example, I can see which cells and the magnitude of which cells, express LHX6, and I can layer that with their Fos activation as a proxy for how stimulated these neurons are. I can then go back and layer that to ask what does CALB1 and CALB2 have to do with that. I'm excited to be able to visualize that data in an interactive format.
As far as the future of my work and the impacts, I’m looking forward to being able to first identify neurons in a solid set that are responsible for tiredness. There is a lot about sleep-promoting neurons that we don't know and that we haven't been able to identify. Going back to the fact that it's been a very heterogenous population, we haven't been able to really pinpoint in a cell-specific manner what these neurons are doing. I believe that this work will help achieve this and provide a library and a catalog with that information.
In terms of the public health impacts, it would be great to definitively characterize which cells are responsible for tiredness and define the molecular mechanisms in which it is regulated. And further, how can we manipulate this pathway to improve the health of those with sleep disorders?
Why is peer-to-peer communication in science important?
PWC: I think it's two-pronged. One, it keeps you informed. It allows scientists to know what's going on and helps you keep up to date with the research in your field. In my opinion, all scientific information should always be sharable. There's always something to learn from your peers and you never know what great insight they might have to offer. Plus, who doesn't want to contribute to general knowledge anyway? That's why we become scientists.
Also, particularly in this climate, it's very important to have peer-to-peer communication in science because we need to discuss how we could do a better job communicating science in a way that's understandable and relevant to the public. Particularly because there's so much widespread misinformation. It's also a part of our job, as scientists, to figure out how to communicate this information beyond our community in the most effective way possible.