CosMx™ SMI - Spatial Multiomics Single-Cell Imaging Platform
The CosMx Spatial Molecular Imager is an FFPE-compatible, single-cell imaging solution that enables researchers to investigate intact tissues through the power of spatial multiomics.
High-speed imaging with strong reproducibility and data quality
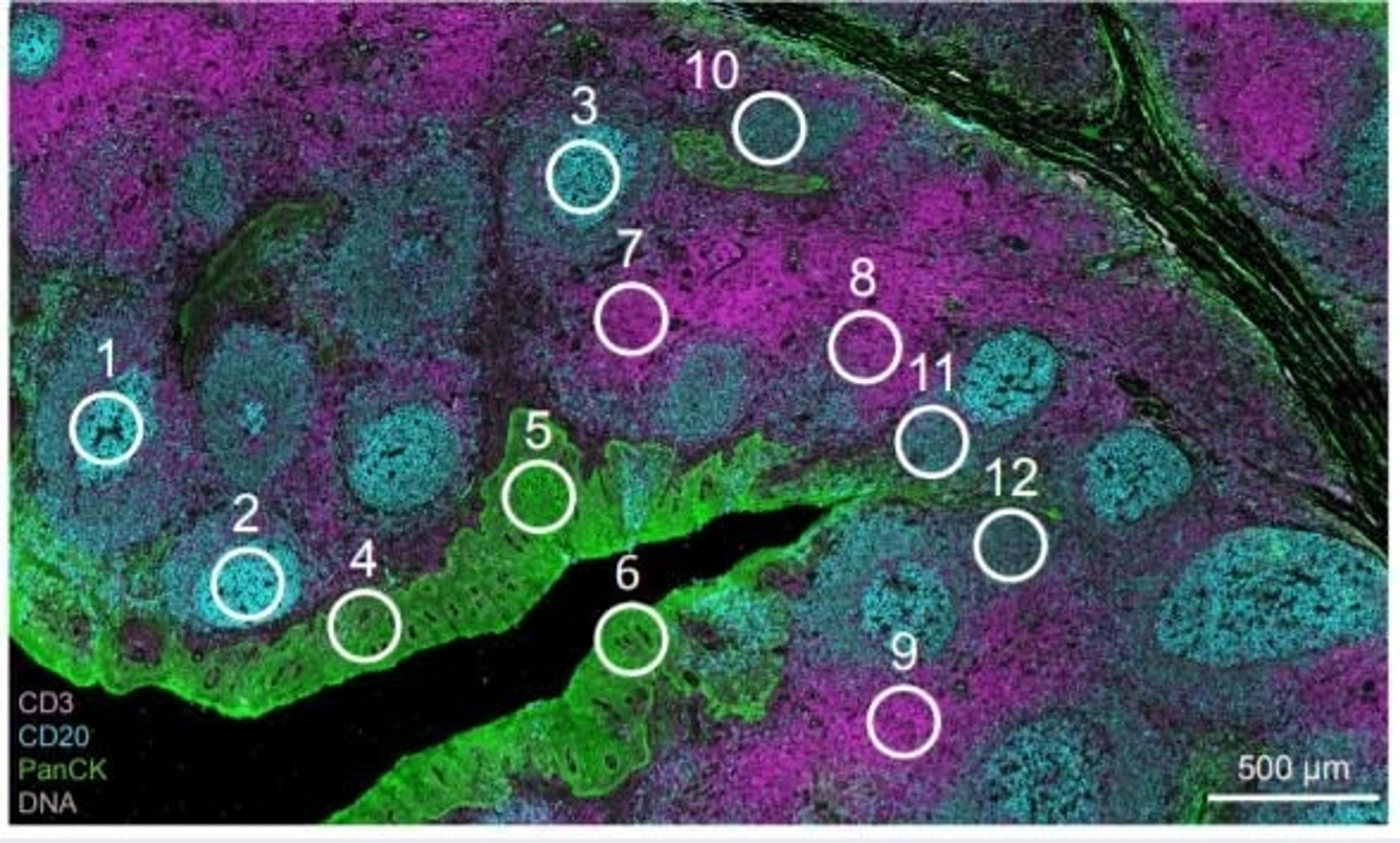
Tumor microenvironment (TME)
The tumor microenvironment (TME) is a complex and dynamic inter and intra-tumor heterogeneity, comprised of an array of cell types. Therefore, it is difficult to recognize the different cell populations using bulk cell approaches. The CosMx Spatial Molecular Imaging (SMI) platform allows for comprehensively map expression profiles of single cells in their spatial environment and extract new insights from a single transcriptomic experiment.
Review Date: 27 Oct 2025 | Bruker Spatial Biology
Demo run result was just amazing.
Establishment of spatial omics core services to all users locally and across university.
The demo run result was just amazing. Can profile tens of thousands of cells from a section within each run. Gene panel options can be improved. Sensitivity and robustness are satisfactory.
Review Date: 29 Sept 2023 | Bruker Spatial Biology
Elevate your single-cell research with Spatial Multiomics Single-cell Imaging Solution
CosMx™ SMI is the first high-plex in situ analysis platform to provide spatial multiomics with formalin-fixed paraffin embedded (FFPE) and fresh frozen (FF) tissue samples at cellular and subcellular resolution. CosMx SMI enables rapid quantification and visualization of up to 1,000 RNA and 100 validated protein analytes. With a tunable workflow from high-plex to high-throughput, CosMx enables discovery research by providing high-plex and unbiased whole-slide analysis, as well as translational research with low-plex, high-throughput biology-driven analysis. It enables cell atlasing, tissue phenotyping, cell-cell interactions, cellular processes, and spatial biomarker signature discovery.
CosMx SMI is an integrated sample-to-result spatial biology solution. The CosMx platform utilizes simple sample preparation based on standard ISH/IHC protocols and offers both custom and pre-validated assay panels for straightforward prep and reliable results. The combination of sensitive cyclic in situ hybridization chemistry, an ultra-high-resolution imaging readout instrument, and an interactive cloud-based data analysis and visualization software makes the data simple to collect and interpret.
Features
- High-plex in situ spatial analysis
- Spatial multiomics with any sample type at cellular and subcellular resolution
- Up to 1,000 RNA and 100 validated protein analytes
Applications
- Cell Atlas and Characterization: Define cell types, cell states, tissue microenvironment phenotypes, and gene expression networks within spatial context.
- Cell-cell Interaction: Understand biological process controlled by ligand-receptor interactions.
- Spatial Biomarkers: Quantify change in gene expression based on treatment and identify single-cell subcellular biomarkers with spatial context.