Mass Profiler Professional Software
Agilent Mass Profiler Professional (MPP) software is a powerful chemometrics platform designed to exploit the high information content of MS data and can be used in any MS-based differential analysis to determine relationships among two or more sample groups and variables.
Agilent Mass Profiler Professional (MPP) software is a powerful chemometrics platform designed to exploit the high information content of MS data and can be used in any MS-based differential analysis to determine relationships among two or more sample groups and variables. MPP provides advanced statistical analysis and visualization tools for GC/MS, LC/MS, CE/MS and ICP-MS data analysis. MPP also integrates smoothly with Agilent MassHunter Workstation, Spectrum Mill and ChemStation software and is the only platform that provides integrated identification/annotation of compounds and integrated pathway analysis for metabolomic and proteomic studies. The system also enables Automated Sample Class Prediction that revolutionizes mass spectrometer-based qualitative analysis of unknown samples in many applications.
Features:
- Allows differential analysis of two or more sample sets from one or multiple MS analysis platforms in a single project.
- Designed for both mass spectrometrists and statisticians, with guided and advanced workflows.
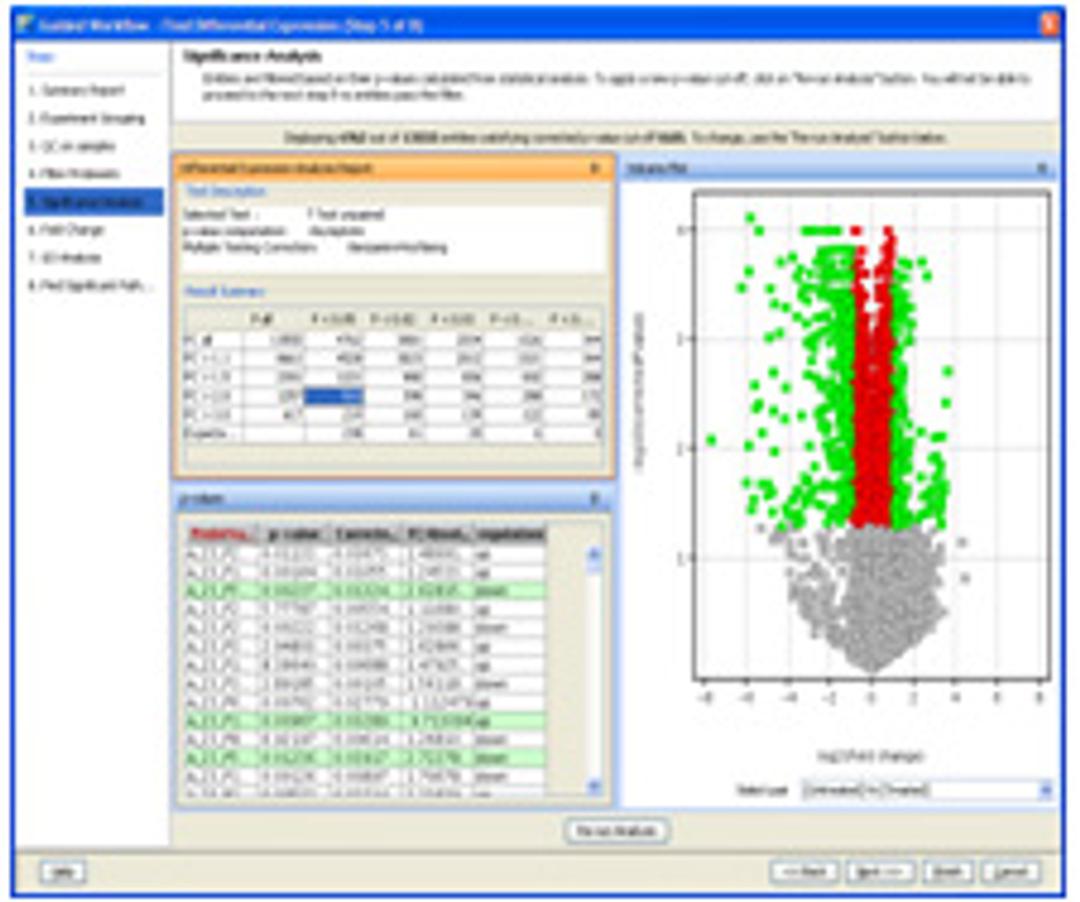
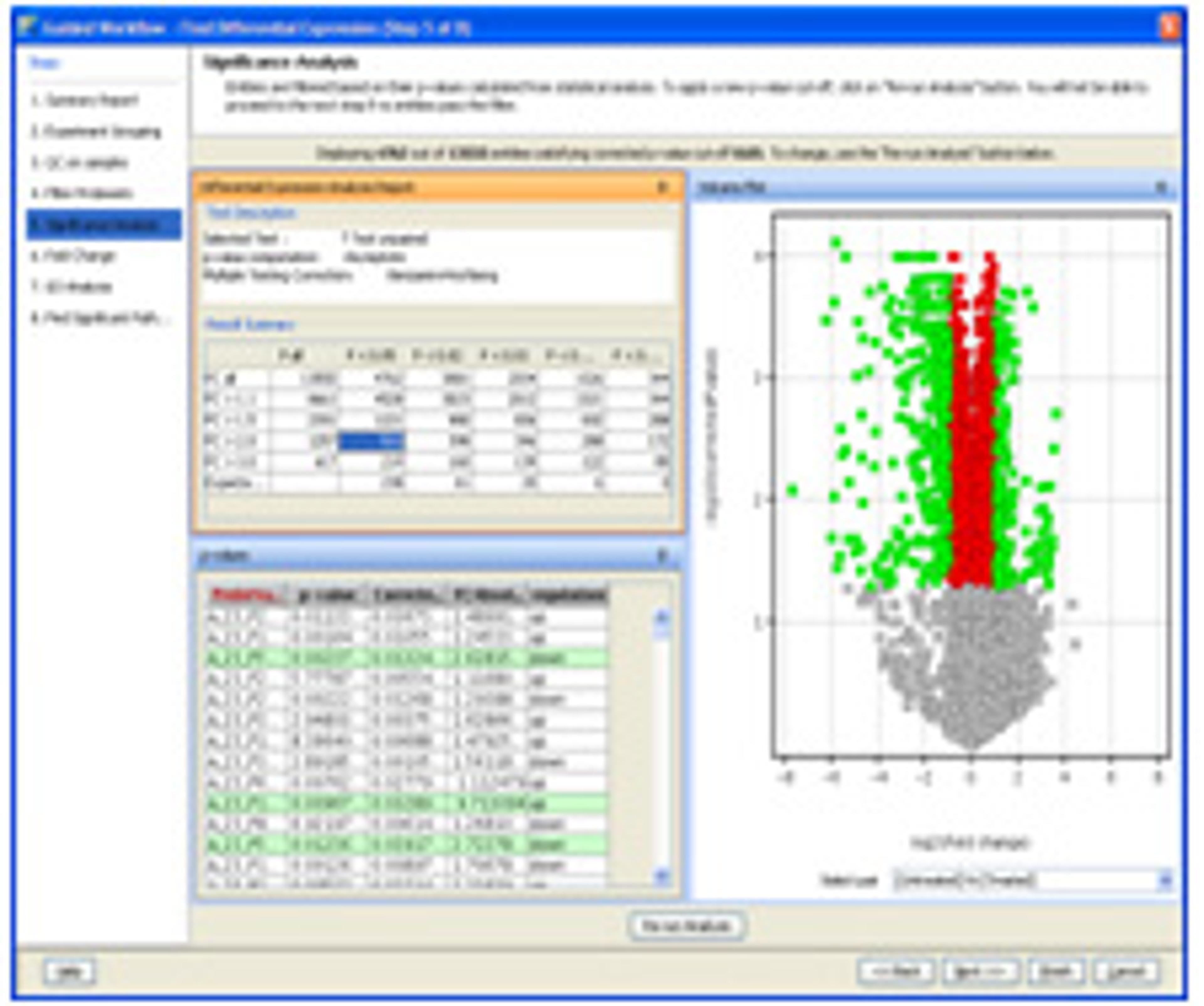
- Provides comprehensive statistical tools including ANOVA, PCA, volcano plots, hierarchical trees, SOMs, QT clustering, and five different methods for class prediction.
- Generates inclusion list for subsequent MS/MS-based analyses to facilitate identification of compounds of interest.
- The ID Browser allows integrated identification and annotation of compound using LC/MS Personal Compound Databases (METLIN, Pesticides, Forensics) and GC/MS libraries (NIST and Agilent Fiehn Metabolomics).
- Allows import and export of data in several formats e.g. to further extend and customize statistical analysis and visualization capabilities using R-Scripts.
- Integrates smoothly with Spectrum Mill to enable TOF/Q-TOF based discovery and QQQ-based verification of protein biomarkers.
- MPP 12 integrated with GeneSpring GX 12 allows the correlation of pathways between MS (metabolomics, proteomics) and micro-array data (transcriptomics) in one project, enabling true integrated biology studies.
- MPP 12 class prediction models from GC/MS, LC/MS, CE/MS and ICP/MS data now can be used with the new, standalone Automated Sample Class Prediction (SCP) software to enable high-throughput routine classification of unknown samples.