Product NewsLife Sciences
Low Input ChIP-seq at Emory University
17 Apr 2017
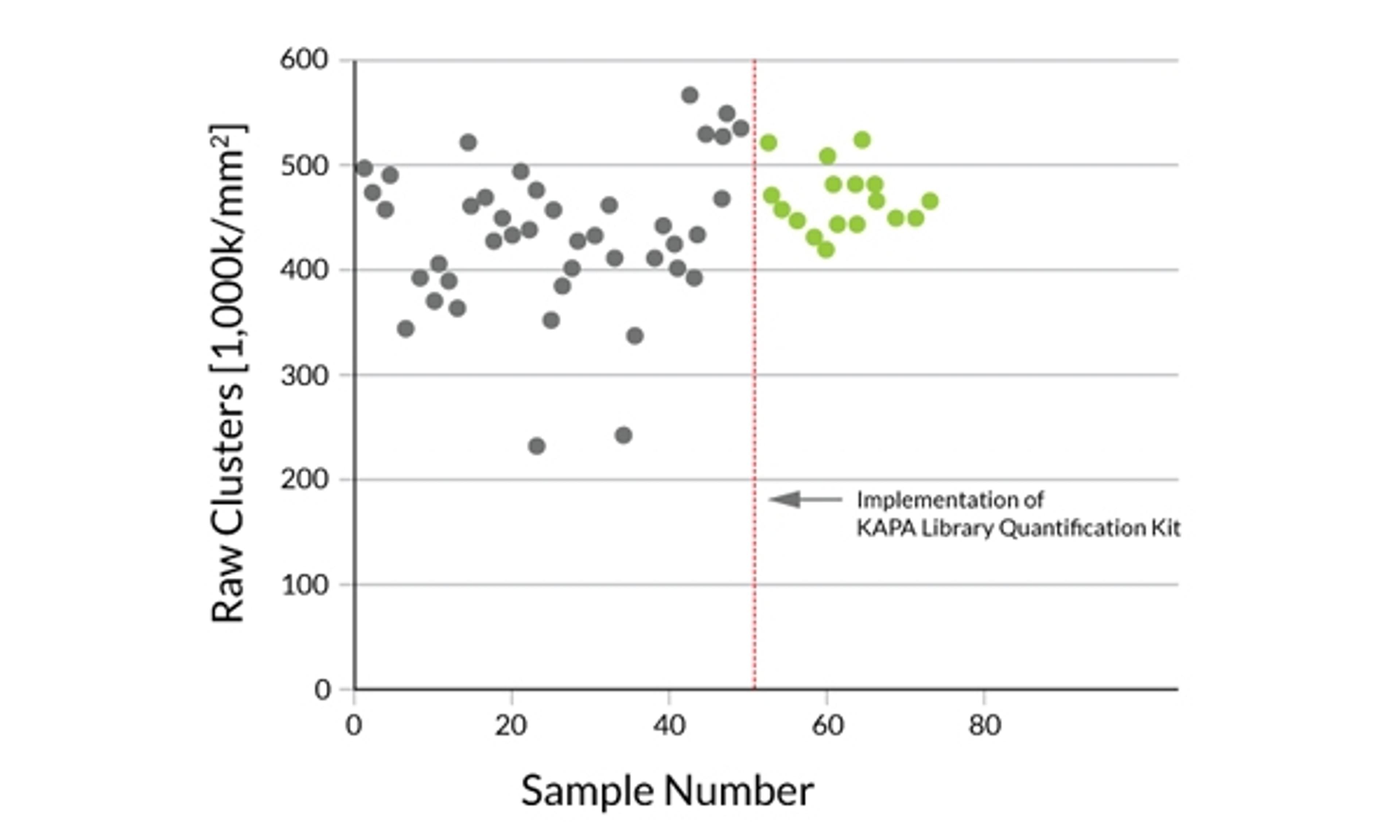
Chromatin Immunoprecipitation sequencing (ChIP-seq) is routinely used to map covalent histone modifications and transcription binding sites. Typically, 1 ng or more of post-ChIP DNA is required to generate a sequencing library. However, as the analysis of epigenomes transitions from cell culture to primary cells and tissues, the amount of post-ChIP DNA is often in the picogram range. Watch this video to hear Dr Chris Scharer of Emory University discuss how to optimize library preparation to overcome this technical limitation.