Efficient chromatin removal in viral vector manufacturing using salt-active nucleases
The first enzymatic solution for complete removal of chromatin at physiological salt conditions and its effect on down stream processing
17 Sept 2024Abstract
The presence of chromatin-associated DNA in crude viral suspensions poses significant challenges for the downstream processing (DSP) of viral vectors. Traditional nucleases derived from Serratia marcescens, such as Benzonase® and Denarase®, exhibit reduced activity under physiological salt conditions (~150 mM NaCl), leading to incomplete chromatin fragmentation.
Recent peer-reviewed studies evaluated the performance of M-SAN HQ, a nuclease specifically optimized for high activity at physiological salt concentrations, in the degradation of chromatin-associated DNA from viral crude harvests.
These publications demonstrate that traditional nucleases not only digest DNA from crude harvests less efficiently than M-SAN HQ, but that the residual DNA is in the form of large chromatin structures causing problems in DSP. These publications show that M-SAN HQ efficiently fragments chromatin to sizes allowing easy removal in the DSP without additional salt. This not only improves the purity of the viral vector but also the potential to enhance DSP efficiency, increase yield, shorten processing times, and reduce costs in manufacturing therapeutic viruses.
Introduction
After using host cells to produce viral vectors or other therapeutic viruses, the primary objective is to isolate, concentrate, and purify the product from cellular constituents and any extraneous materials introduced during the process. Host cells are subjected to significant stress to maximize viral vector production, resulting in cell death and the release of cellular components into the media that must be removed during the DSP.
One of the critical challenges in purification is the DNA released is in the form of dense chromatin rather than naked DNA. Due to its complex attraction profile and large size, chromatin poses significant challenges in the purification process.
Traditionally, endonucleases used for the removal of DNA during the production of viral vectors originate from S. marcescens.This nuclease is available from several vendors under different names, such as Benzonase® (Merck) and Denarase® (c-LEcta). Typically, the nuclease is added to the cell lysate or to the supernatant of spent cell media before DSP to simplify the purification process. However, the activity of these nucleases drops by nearly half under physiological conditions (cell media) and becomes almost inactive at higher salt concentrations (~500 mM NaCl), which are often used to break down aggregates formed by polar interactions between virus particles, chromatin and other cell debris.
To address these limitations, ArcticZymes developed two nucleases, each tailored to the bioprocessing conditions used for viral vector production. The first, M-SAN HQ is specifically designed for the physiological salt conditions found in cell media (~150 mM NaCl) while the second, SAN HQ is optimized for use at high salt concentrations (500 mM and above).
Although high salt concentrations are effective for manufacturing robust viral vectors like adeno-associated viruses (AAVs) and adenoviruses, they are unsuitable for more sensitive vectors, such as lentiviruses.
In this report, we show that M-SAN HQ, unlike Benzonase and Denarase, efficiently removes complex DNA in the form of chromatin at physiological conditions, thereby significantly improving the manufacturing process of fragile viral vectors, such as lentiviruses.
The landscape of salt-active nucleases
The first salt-active nucleases for biomanufacturing, SAN HQ and M-SAN HQ, were pioneered by ArcticZymes starting in 2017. These nucleases were designed to address the specific needs of bioprocessing at both physiological and high salt conditions. M-SAN HQ is optimized for use in cell media (125-200 mM NaCl), while SAN HQ is engineered for high salt concentrations (≥400 mM NaCl).
Recognizing the value of salt-active nucleases in biomanufacturing, Merck and c-LEcta recently introduced nucleases targeting the disaggregating salt range. However, neither has been specifically optimized for performance at physiological salt conditions, found in cell media. Instead, c-LEcta appears to be targeting broader functionality across a wide salt range, rather than focusing on the specific salt conditions most commonly used.
The comparative efficacy of these new nucleases and ArcticZymes’ products was evaluated using a model system that measured DNA digestion at the pH and temperature conditions typical of bioprocessing. Given that nucleases generally require higher MgCl2 concentrations than those in cell media for optimal performance, 5 mM MgCl2 was added.
The results indicate that M-SAN HQ significantly outperforms rivals at physiological salt levels.
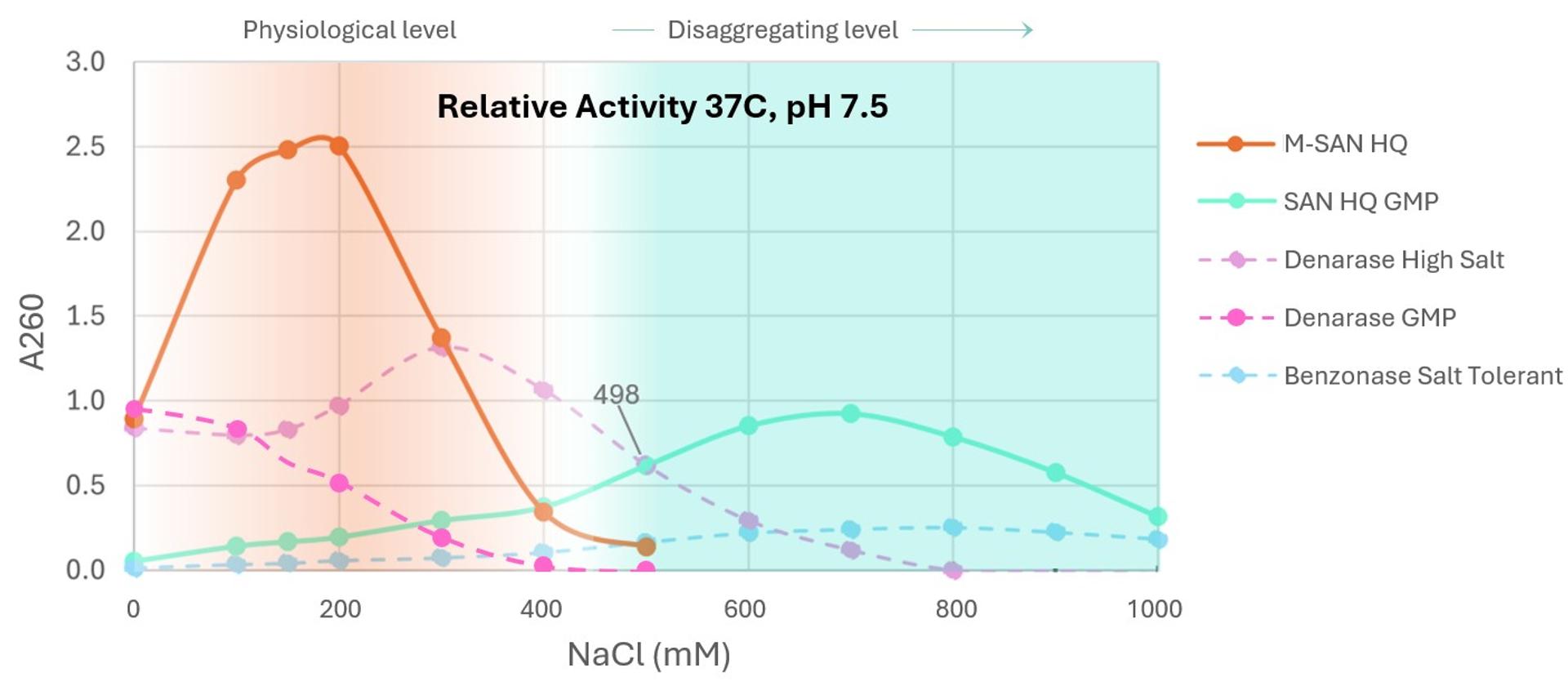
Figure 1: At physiological salt (corresponding to 135 – 150 mM NaCl), M-SAN HQ shows higher activity than other commercially available nucleases. The reactions were incubated 20 min at 37°C with 0.2 mg/ml Calf Thymus DNA, 5 mM MgCl2, 25 mM Tris-HCl (pH 7.5) @ 37°C and various salt concentrations. The reactions were stopped by adding perchloric acid, incubated on ice for 20 min, centrifuged and measured at Abs260.
Comparison: removal of chromatin-associated DNA in viral suspensions
Recent publications have demonstrated that M-SAN HQ is superior for fragmenting chromatin in viral vector manufacturing at typical cell media conditions. Pagallies et al. compared the performance of M-SAN HQ and Denarase (recommended by c-LEcta for use at NaCl levels below 200 mM) during the scale-up of manufacturing fragile, large, and enveloped Orf viruses in suspension cell culture1. Pagallies’et al. showed that, compared to Denarase, M-SAN HQ removed significantly more DNA from crude harvest. Using M-SAN HQ significantly enhances the efficiency of DNA digestion at 37°C, reducing both the digestion time and the amount of nuclease required, while also improving overall DNA removal (Figure 2).
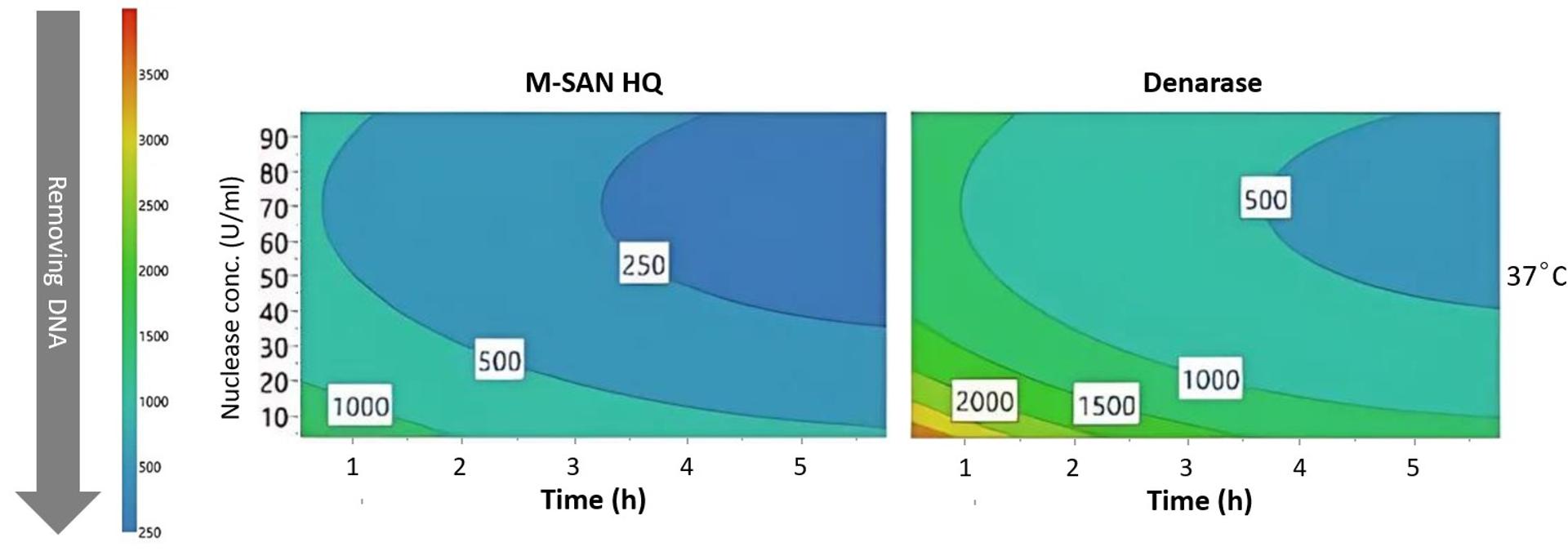
Figure 2. 4D contour plot of the DNA concentrations (ng/ml) following nuclease digestion of a crude harvest of Orf virus using varying nuclease concentrations and incubation times. Digestion was conducted at 37°C with 150 mM NaCl and MgCl₂ was added to a concentration of 2 mM for Denarase, and 10 mM for M-SAN HQ. Figure adapted from Pagallies et al1.
The data from Pagallies et al. suggest that some DNA present in the crude harvest is not degraded by Denarase but can be effectively removed by M-SAN HQ. The critical question is the state of the remaining DNA and its potential impact on DSP.
Data from Aguilar et al. at the Austrian Centre of Industrial Biotechnology (ACIB) provide further insights2. Clarified cell culture supernatants containing HIV-1 gag virus-like particles (VLPs) were treated with Benzonase (which performs identically to Denarase), pre-processed using CaptoTM Core 700 for flow-through chromatography, and further purified with CaptoTM Heparin affinity chromatography. Despite this comprehensive treatment, significant amounts of chromatin fragments, even larger than the VLPs themselves, were detected in the purified product using Western blots targeting Histone H3 and Transmission Electron Microscopy (TEM), as shown in Figure 3.
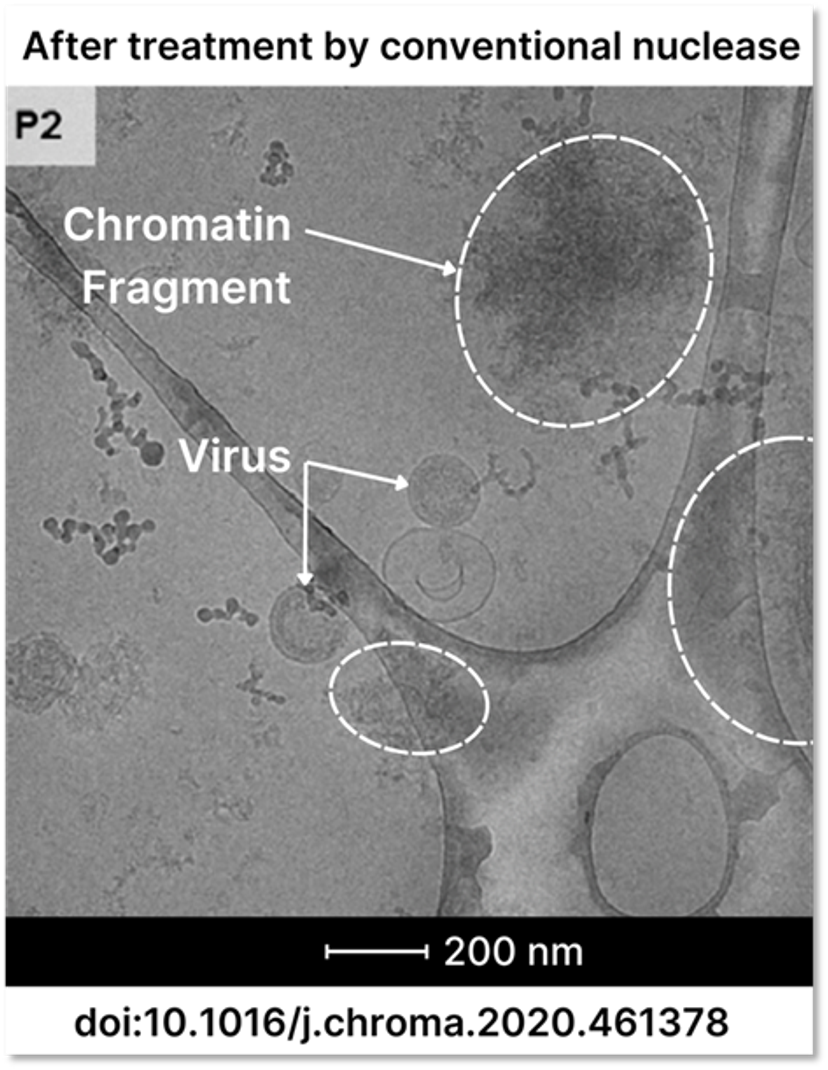
Figure 3. Clarified cell culture supernatant was treated with Benzonase, then preprocessed using flow through chromatography and further purified using a Capto Heparin column. Eluates from the Capto Heparin step were studied using Transmission Electron Microscopy and showed both virus particles as well as large chromatin structures. Figure adapted from Aguilar et al2
Researchers at ACIB then tested whether salt-active nucleases could better remove chromatin from recombinant measles virus3. In Mayer et al. (2023), the measles virus was chosen as a candidate due to its tolerance to high salt concentrations, allowing for testing of DNA removal at both physiological cell-media conditions and disaggregating salt levels. Mayer et al. compared the performance of Benzonase, Denarase, and M-SAN HQ at physiological salt levels and SAN HQ at high salt levels. After DNA digestion of the cell culture supernatants, the material was passed over a Capto Core 700 column, and the Histone H3 protein presence was evaluated using Western blot.
The findings supported Aguilar et al. (2020), show that neither Denarase nor Benzonase could sufficiently fragment chromatin. However, both M-SAN HQ and SAN HQ removed chromatin to levels undetectable by Western blot at its respective ideal salt concentration (Figure 4). While it was expected that the combination of high salt and digestion with SAN HQ would efficiently fragment chromatin, it was unexpected that a nuclease could achieve the same without the contribution of high concentrations of salt. The data clearly showed that M-SAN HQ efficiently degrades chromatin into small fragments at physiological conditions, simplifying its separation from viral vectors based on size.
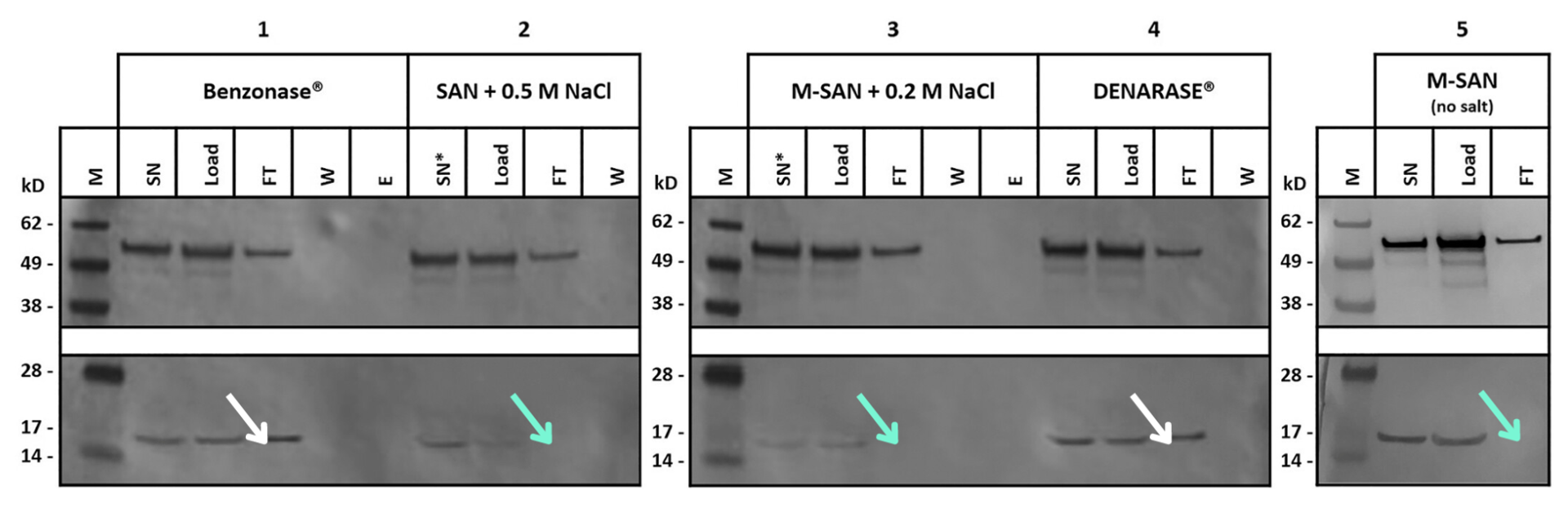
Figure 4: Western blots detecting viral vectors (nucleoprotein, top) and chromatin (histone H3 protein, bottom). SN, supernatant (clarified virus suspension); SN*, high salt supernatant; Load, supernatant after nuclease digestion; FT, flow-through; W, wash; E, elution. Mayer et al3
Manufacturers of fragile viral vectors have historically faced challenges with chromatin remnants following nuclease digestion, often without full awareness of this issue. However, the application of M-SAN HQ for treating crude viral suspensions under physiological conditions enable complete chromatin removal. This advancement opens numerous possibilities for improved and simplified DSP by eliminating one of the most problematic contaminants before DSP even begins.
Potential benefits include shortened workflows, reduced DNA contamination, lower costs, and higher yields. Further investigations are necessary to fully understand how the appropriate nuclease can enhance the large-scale manufacture of fragile viral vectors.
Conclusion
Peer-reviewed studies have demonstrated that M-SAN HQ excels in removing chromatin-associated DNA at physiological salt conditions, significantly outperforming traditional nucleases like Benzonase® and Denarase®. These findings show that M-SAN HQ efficiently fragments chromatin, simplifying DSP, improving viral vector purity, and eliminating the need for high salt conditions. This is particularly advantageous for fragile viral vectors such as lentiviruses, where high concentrations of salt can be detrimental.
M-SAN HQ achieves the same level of chromatin removal at physiological salt levels that was previously only possible with high salt concentrations. This allows reduced processing times, and lowers cost in viral vector manufacturing. Together with SAN HQ for high salt conditions, ArcticZymes offers solutions specifically designed to meet the distinct needs of biomanufacturers. The consistent results from these publications highlight the value of M-SAN HQ in optimizing DSP and advancing viral vector production.
References
1. Pagallies, F., Labisch, J. J., Wronska, M., Pflanz, K. & Amann, R. Efficient and scalable clarification of Orf virus from HEK suspension for vaccine development. Vaccine X 18, 100474 (2024).
2. Pereira Aguilar, P. et al. Capture and purification of Human Immunodeficiency Virus-1 virus-like particles: Convective media vs porous beads. J. Chromatogr. A 1627, 461378 (2020).
3. Mayer, V. et al. Removal of chromatin by salt‐tolerant endonucleases for production of recombinant measles virus. Biotechnol. Prog. 39, e3342 (2023).

