Using in situ RNAscope analysis to visualize spatial gene expression in plants
Learn how RNAscope technology was adapted to study plant tissues and gain insights into plant immunity
26 Jul 2020

Conventional RNA analysis using in situ hybridization (ISH) has lacked sensitivity and robustness, while qPCR and microarray profiling that involve RNA extraction have deprived spatial and morphological information. The introduction of RNAscope in 2011 resolved these RNA-based challenges by introducing a novel RNA ISH that allows simultaneous signal amplification and background suppression to achieve visualization while preserving tissue morphology.
In this article, we speak with Dr. Shyam Solanki, of Washington State University, Pullman, WA, USA, who recently developed and published a method to study in situ RNA expression in plant cells, an underexplored tissue type for RNAscope applications. A postdoctoral researcher in Dr. Robert Brueggeman’s lab, Solanki explains why it’s important to study plant immune responses to pathogens and how he adapted the RNAscope technology to develop a plant-based technique.
Disease resistance genes
Plant infections caused by bacteria, viruses or fungi can result in yield loss in agriculture. In plants, disease resistance is conferred by cell surface and cytoplasmic receptors and the majority of them belong to the nucleotide-binding leucine-rich repeat (NLR) proteins, the intracellular immune receptors that recognize pathogen proteins. “Stem rust, a fungal pathogen, is a big problem in wheat and barley plants. Recently, there have been reports of virulent strains of this pathogen spreading across Europe and other parts of Asia,” says Solanki. “We study NLR genes that provide sensing patterns for the stem rust pathogens.”
The overarching goal of the Brueggeman lab is to develop synthetic plant immune receptors that can provide a broad spectrum of resistance to pathogens without resulting in yield penalties for the agricultural industry.
In studying the disease resistance genes, analyzing RNA expression in situ can provide deeper insights into gene functionality and reveal distribution patterns within a cell. However, RNAscope, widely used for in situ RNA analysis, has mostly been applied to mammalian cells and tissues.
Developing a method
The decision to extend RNAscope to plant biology stemmed out of pure curiosity. Solanki recalls: “When I was working at an advanced imaging center at the North Dakota State University, I noticed a brand-new piece of equipment on the working bench. I learned that it was an RNAscope hybridization oven that was meant to be used in the future. I found the term ‘RNAscope’ rather catchy. When I looked it up, I learned about its fantastic and impressive RNA detection capabilities in tissue sections. So, I told myself: Let’s test this on plant tissues.” Collaborating with the director of the advance imaging center, Dr. Pawel Borowicz, Dr. Solanki and Dr. Brueggeman came up with the idea of validating candidate genes in the barley plant using RNAscope.
To get reliable and reproducible results, every assay begins with a robust method validation step. Solanki and colleague Dr. Gazala Ameen faced a unique challenge in that the RNAscope protocols developed for mouse and bovine tissues could not be directly applied to plant tissues due to their tough cell wall morphology, making sectioning and visualization more difficult. “I decided to learn and test out the whole method using mouse and bovine tissues first. This gave me a good indication of whether or not I was doing it right,” says Solanki. “I eventually started working with the plant system, tweaking the protocol one step at a time. In the process, I would test sample collection methods, increase or decrease probe incubation times, test different chemical concentrations to finally figure out what works best for plant tissues.”
Solanki then validated this assay with two different genes, HvGAPDH and Rpg1. “GAPDH is a classically known housekeeping gene, and Rpg1 is a well-characterized resistant protein in barley against stem rust,” explains Solanki. “We designed the probes using help from the ACD Bio probe designing team. Once we had the probes, we optimized the protocol. The method worked wonderfully! After we optimized it on plant tissues, it was very straightforward and easy to use.”
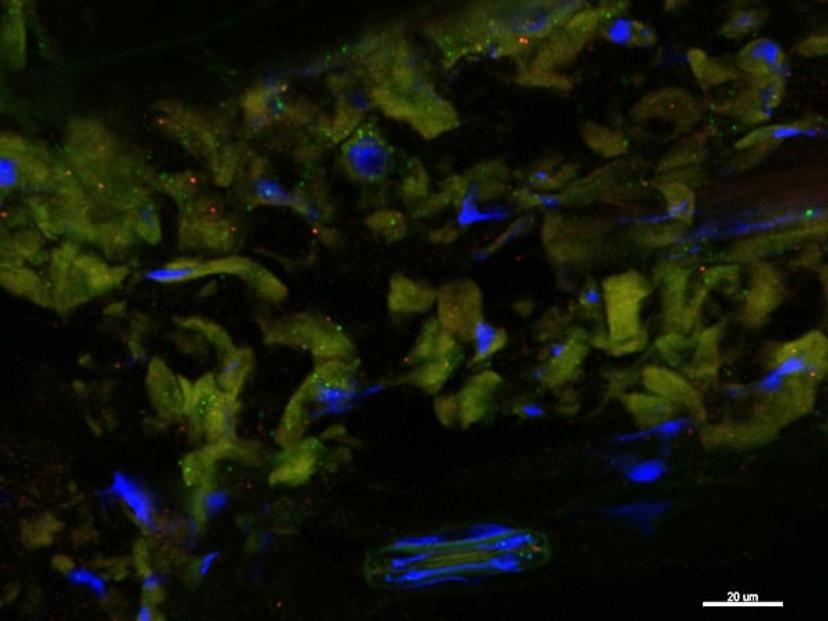
Published results
Results of the method developed by Solanki and the team were recently published in Plant Methods (https://doi.org/10.1186/s13007-020-00614-4). Summarizing the significance of the paper’s findings Solanki explains: “Rpg1 is a gene which has been previously characterized. It was shown to have low expression, except in the epidermal tissue sections. These previous studies used epidermal peel assays and qPCRs. So, the actual site of expression remained unknown.”
“Using the RNAscope assay, we were able to specifically pinpoint the expression of the Rpg1 gene. We found that it is located near the stomatal guard cells,” Solanki adds. “We know that the stem rust pathogen infects plants by going through the stomates. With these findings, we are now able to hypothesize that Rpg1 is expressed at higher levels near the stomate where it possibly stops the entry of pathogens.”
The ability to analyze gene expression in situ using RNAscope makes it possible to visualize signals while preserving the tissue morphology that would be otherwise lost with conventional RNA extraction methods. Solanki and the other members of the Brueggeman lab are now using the RNAscope to characterize and dissect other immune pathways in barley, wheat, and Arabidopsis, focusing on stomatal and kinase genes. Solanki continues: “We recently started working on carbon nanotube-mediated transient expression system so we can rapidly test our hypothesis and study candidate genes in barley and Arabidopsis. We will be using RNAscope to confirm transgene expression patterns.”
Future food security
Studying genes involved in plant immunity can have direct implications on agriculture and the food economy. “The future of plant immune studies lies in engineered plant immune receptors,” says Solanki. “By fine-tuning their pathogen sensing capability, we can integrate engineered receptors into agriculturally important plants, providing them with better resistance against pathogens.” In addition to disease resistance, synthetic plant receptors can also be used to provide food security as the global climatic patterns undergo unprecedented changes. Fluctuating temperatures and unpredictable rainfalls can pose increased pathogen risks, making plants more susceptible to infection. Using engineered immune receptors that help build resistance to new pathogen varieties can protect food plants. “By examining plant immune genes, devising synthetic immune receptors, and testing them on different plants, we can provide stronger immunity to our plant systems,” concludes Solanki.
Here more from the RNA ISH experts:
- How spatial genomics is revolutionizing our understanding of disease
- Early detection of prostate cancer through game-changing molecular markers
- Plus, download your free application eBook on RNA ISH single-cell analysis

