Early detection of prostate cancer through game-changing molecular biomarkers
Dr. Nallasivam Palanisamy, Henry Ford Cancer Institute, discusses his discovery of the first pseudogene-associated recurrent gene fusion in cancer and the potential for this robust biomarker for application beyond prostate cancer
2 Aug 2020

For men around the world, prostate cancer is the most common cancer type and the second most common cause of cancer deaths. However, despite its prevalence and morbidity, the diagnosis, treatment, and management of prostate cancer remains a challenge for clinicians and translational researchers.
In this exclusive SelectScience® interview, we hear from Dr. Nallasivam Palanisamy, Associate Scientist at the Henry Ford Cancer Institute, about his work to identify new prostate-cancer-specific molecular targets that can be used for early detection and development of treatments, including the groundbreaking discovery of pseudogene-associated recurrent gene fusions that promise more robust, rapid, and non-invasive cancer diagnosis.
The functional role of gene fusions and pseudogenes in cancer
Methods of gene fusion discovery
1. Karyotyping chromosomes, e.g. BCR/ABL1 in myeloid leukemia
2. Microarray technology, e.g. TMPRSS2-ERG in prostate cancer
3. NGS technology, e.g. the KLK4-KLKP1 in prostate-cancer
Prostate specific antigen (PSA) screening is a commonly performed test used to diagnose prostate abnormalities, including inflammation and cancer. Generally, a raised PSA level is considered to be a sign of prostate cancer. However, this is not always the case and the test is notorious for false positives and negatives. Despite this, more robust prostate-cancer-specific markers have yet to be identified.
One increasingly promising area in cancer biomarker research is the identification of gene fusions – hybrid genes formed by the chromosomal rearrangement and recombination of two previously independent genes. Using traditional methods, gene fusion mutations have been found in lymphomas and leukemias and, in 2005, the University of Michigan reported the first recurrent gene fusion present in 50% of prostate cancer cases – the TMPRSS2-ERG gene fusion.
In 2007, Palanisamy looked to new next-generation transcriptome sequencing (NGS) technology in order to sequence the remaining 50% of prostate samples that were negative for this TMPRSS2-ERG gene fusion. This led to his group’s discovery of the RAF kinase gene fusions, SLC45A3–BRAF and ESRP1–RAF1. The key benefit of these gene fusions is that RAF kinases make much better therapeutic targets than the classical ETS-family transcription factors and are already treatable with existing therapeutics. “Only a small number of prostate cancers have this gene fusion, up to 2%, but this is now an extra 2% of cases that will now benefit from treatment,” says Palanisamy. “I’m now continuing with this research as there is still a large subset of cases where we don’t know what is going on yet.”
Non-invasive test for prostate cancer
An additional benefit to the KLK4-KLKP1 gene fusion is that the resultant protein is detectable through patient urine samples, which allows for non-invasive detection of prostate cancer, before biopsy collection.
Palanisamy’s group amassed a large amount of NGS data which provided them the opportunity to look at the huge non-coding RNA landscape, particularly pseudogenes. Through this research, Palanisamy’s group was able to identify 8 pseudogenes that were expressed in prostate cancer but not in normal prostate tissue. Through the study of these pseudogenes, Palanisamy discovered a new gene fusion marker, formed via the fusion of KLK4, a well-known protein-coding gene, and KLKP1, a non-coding pseudogene thought to be non-functional or junk DNA. “This the first time in any cancer that a recurrent gene fusion has been found that is associated with a pseudogene,” Palanisamy explains, “the KLK4-KLKP1 fusion protein is prostate-cancer-specific and is present in around 32% of prostate cancer cases.”
Simultaneous marker evaluation for multifocal cancer
Alongside the ever-increasing need for alternative biomarkers to PSA, there is also the drive to be able to handle metastasis. 90% of prostate cancer cases are multifocal, meaning they present with more than one tumor. Traditionally, clinicians thought that the index tumor – the largest and most highly differentiated ‘dominant’ tumor – drove the biology of the disease. However, studies have shown that it is not always the index tumor that establishes metastasis and even small secondary foci can contribute to this process. This, along with the complex genetic diversity of tumors, means that one single marker is not enough for reliable identification of cancer at diagnosis. Therefore, it is essential for researchers to be able to screen for all likely biomarkers to get the full picture.
Conventionally, researchers used immunohistochemistry (IHC) to try to assess the tissue-level expression of the genes. However, whilst there are well-characterized, cancer-specific antibodies for ERG, there aren’t any guaranteed for other associated transcription factors, and it is impossible to produce antibodies for pseudogenes. To overcome this challenge and assess the inter- and intra-tumor molecular heterogeneity in prostate biopsy samples, Palanisamy’s group developed a combined IHC and RNA in situ hybridization (ISH) method utilizing Advanced Cell Diagnostics’ RNAscope technology.
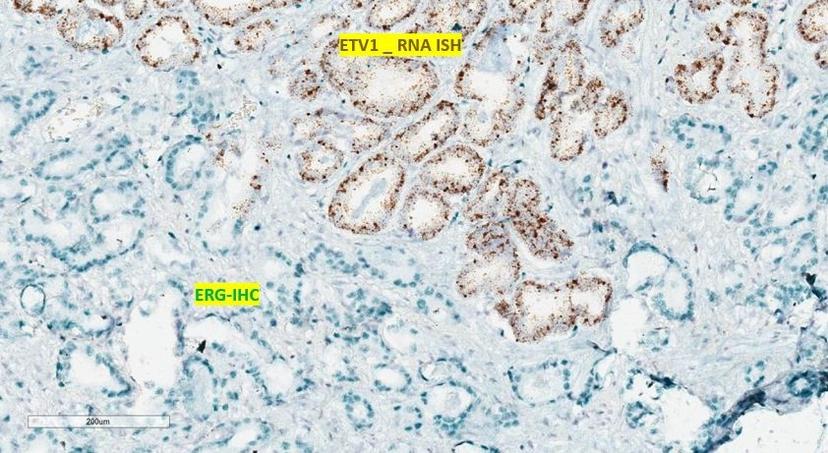
“The RNAscope is a very powerful and unique tool because, as these pseudogenes are non-coding RNA and therefore do not express proteins, you cannot develop any antibodies for them,” Palanisamy explains, “as a result, you can’t exclusively use IHC to study their expression, which is where RNA ISH is invaluable.”
Another noted benefit of the RNAscope technology, for prostate cancer research in particular, is the multiplexing capabilities. With prostate studies, you are limited by the availability of tissue, but with the RNAscope you can perform RNA ISH and then IHC on the same slide. “We have multiple markers to evaluate,” Palanisamy notes, “and we can now look at up to four markers simultaneously to assess the tissue-level expression with the additional information on spatial distribution throughout the prostate, whilst preserving tissue morphology.”
Palanisamy goes on to clarify that: “This is not only limited to prostate cancer, for anyone who wanted to study any gene expression at the tissue level, even if they don’t have established antibodies, RNAscope is the best method.”
RNAscope is a very powerful and unique tool.
Dr. Nallasivam Palanisamy Henry Ford Cancer Institute
Widespread applications and remaining cancer subtypes
Future work
The availability of patient resources from disparate population groups at the institute offers the unique advantage of being able to conduct research concerning racial disparity in prostate cancer. With multiplexed IHC and RNA ISH, Palanisamy’s group has been able to evaluate 1,000 whole-mount radical prostatectomy tissues in an exciting new project, funded by the U.S. Department of Defence (DOD), to uncover why African American men have higher prostate cancer mortality rates.
Palanisamy explains that deeper research always leads to more questions than answers, especially when it comes to cancer: “We have come a long way in our understanding of the genetic underpinnings in prostate cancer, but new approaches help to unravel the ever-increasing complexity of the disease rather than leading us to any favorable solutions.”
Palanisamy’s primary focus is on prostate cancer, but he has additional involvement in several collaborative projects concerning melanoma, kidney cancer, and bladder cancer. As the existing biomarkers are not generally specific to one type of cancer, those identified in one can be applied for use against several others. Palanisamy hopes that this use of NGS technology to analyze the genome-wide expression will pave the way for the discovery of pseudogene-associated biomarkers in other cancers.
“The markers identified thus far only account for around ~60% of prostate cancer subtypes, meaning there are still ~40% of cases where the genetic basis is not known,” Palanisamy concludes. “The need for new molecular markers remains, especially to help distinguish between indolent and aggressive cancers, and to identify any reasons for racial disparity.”
References
- Clonal evaluation of prostate cancer molecular heterogeneity in biopsy samples by dual immunohistochemistry and dual RNA in situ hybridization
- Evaluation of tissue PCA3 expression in prostate cancer by RNA in situ hybridization
- MDA PCa PDX captures the molecular landscape of prostate cancer and facilitates marker-driven therapy development
Here more from the RNA ISH experts:

