CloneTracker XP™ Lentiviral Expressed Barcode Libraries
Easily label cells with an RNA-expressed barcode readily detected by RNA-Seq analysis

The supplier does not provide quotations for this product through SelectScience. You can search for similar products in our Product Directory.
Single-cell RNA-Seq analysis is an important tool to analyze individual cell responses. However, important genes and pathways may not be active in all cells. Certain responses may only occur in a portion of cells, or different groups of cells can manifest divergent phenotypes. One way to address this challenge is to label individual cells with uniquely identifiable, expressed barcodes. In this way, effects and responses of each of the clonal progeny from this initial barcoded population can be assessed, and data correlating to how experimental conditions, differentiation, or developmental processes may affect distinct groups of cells in a population can be extracted.
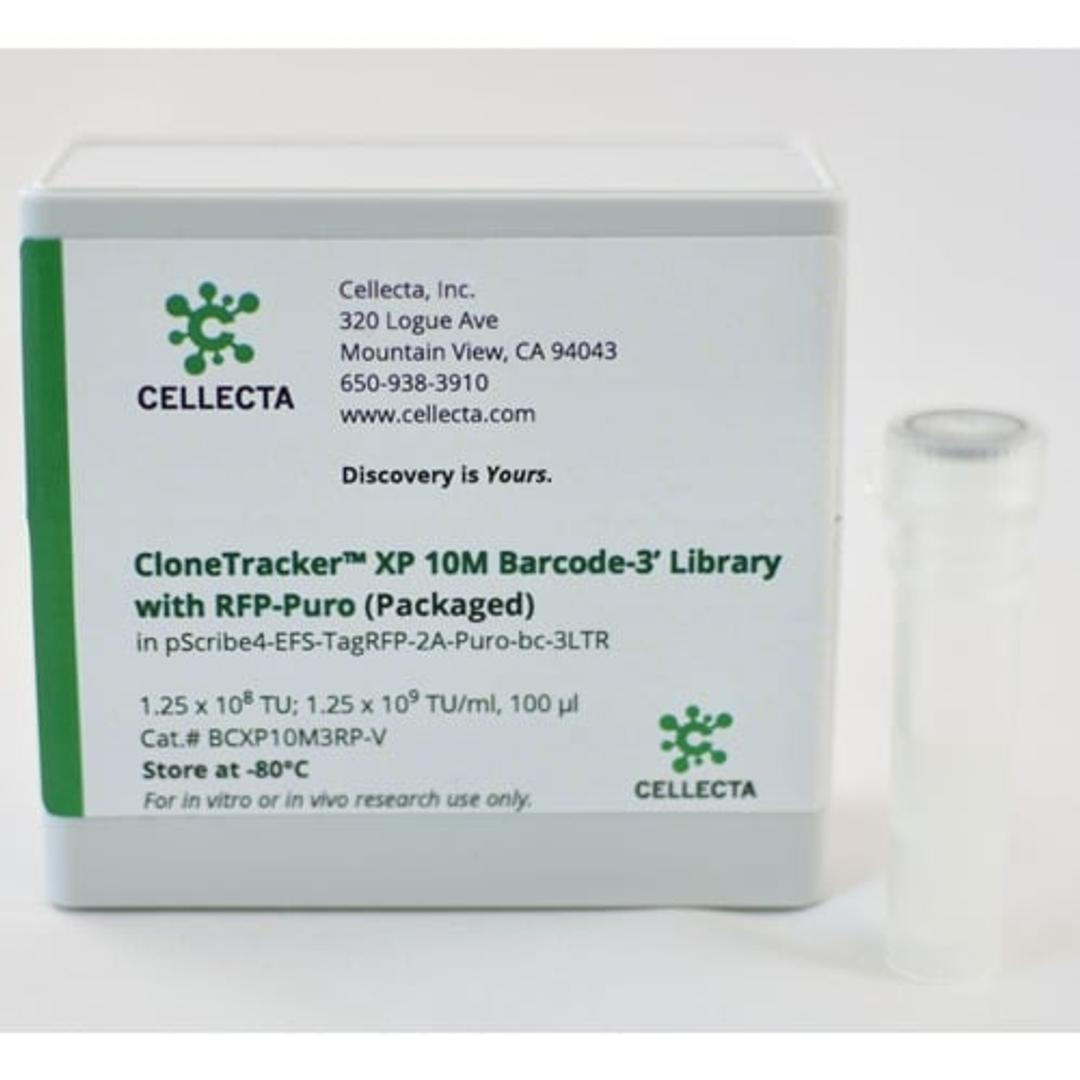
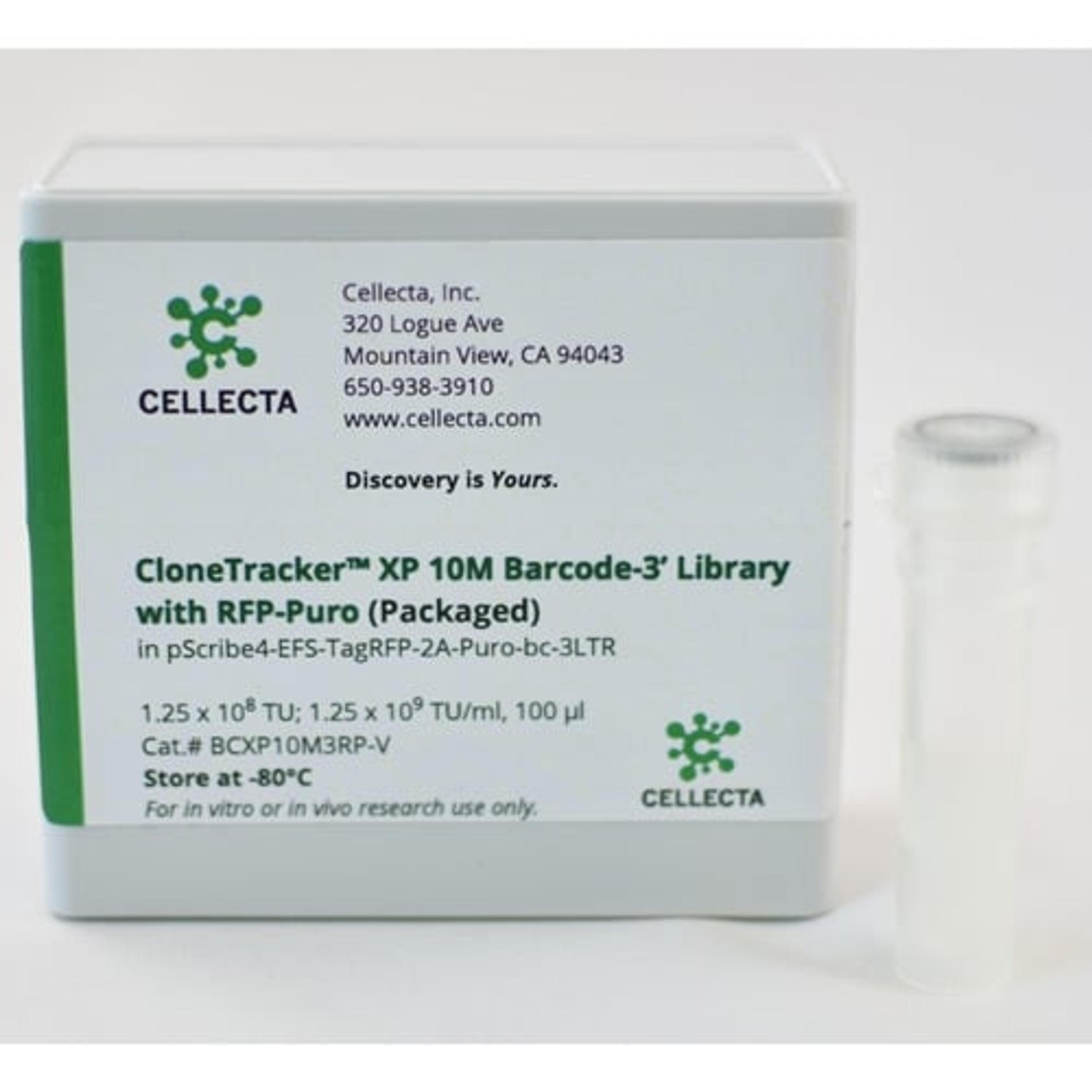
To detect cell-specific barcodes in conjunction with gene activation or pathway analysis, however, barcodes need to be expressed on the mRNA. For this purpose, Cellecta’s CloneTracker XP libraries incorporate barcodes into the 3′- or 5′-UTR of an RNA transcript. These ready-to-use expressed barcode libraries enable convenient labeling of up to 1 million individual cells with unique barcodes that are detected in both the genomic DNA and expressed RNA fractions.
In conjunction with single-cell RNA-Seq analysis, the expressed barcodes may be used to identify expression profiles and activated genes in different cells so that distinct clonal populations of cells from a single progenitor, as well as subpopulations of cells with distinct pathway activations or expression profiles, can be readily identified.
CloneTracker XP 10M Barcode Libraries can be used to label approximately 1M cells with individual unique barcodes detectable by targeted amplification followed by DNA sequencing or RNA sequencing. These libraries are available in configurations where the barcode is located at the 3′ end of the transcript (as shown above) so it is compatible with oligo-dT cDNA synthesis protocols, or a 5′ barcode option that is compatible with SMART-based cDNA synthesis.
CloneTracker XP 1M Barcode Libraries can be used to label a few hundred thousand cells with individual unique barcodes detectable by targeted amplification followed by DNA sequencing or RNA sequencing. We offer multiple pools of these smaller barcode libraries–each with non-overlapping sets of barcodes–so that cells in several populations can be barcoded with differentially identifiable sets of sequences. Changes in the barcode frequency of different cell populations can then be independently analyzed when they are cultured together. Up to 10 different pools of non-overlapping barcode sets are available.