Hyperphage M13 K07ΔpIII
Provides helper phage function in packaging a common phage display phagemid. Infection of bacteria via pIII.

The supplier does not provide quotations for this product through SelectScience. You can search for similar products in our Product Directory.
An Effective Tool for the Isolation of Recombinant Antibodies, Proteins or Peptides from Hyperphage-Packed Libraries.
Advantages
- increases panning efficiency
- allows panning with reduced amount of panning antigen
- identifies high and low affinity binders
Introduction
A helper phage technology ("Hyperphage System") was developed by Rondot et al. (Nature Biotechnology 19:75-81, 2001). The Hyperphage System allows to improve antibody presentation in phage display by increasing the number of antibodies displayed per phage particle (up to 5 vs. 0.01) and thereby the system offers great advantage in the fields of functional gene analysis and proteomics. Panning of phages can be performed with small amounts of antigen and higher efficiency. For example, the application of universal libraries for antibody isolation can be improved by employing panning of hyperphage-packed libraries on blots of protein spots after 2-dimensional gel electrophoresis.
More Applications
Large numbers of open reading frames (ORFs) can be analyzed by panning against synthetic membrane- bound peptide epitopes. In cancer research, the hyperphage-packed library could be a tool to discover new tumour markers by panning against cellular surfaces.
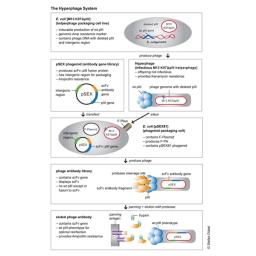
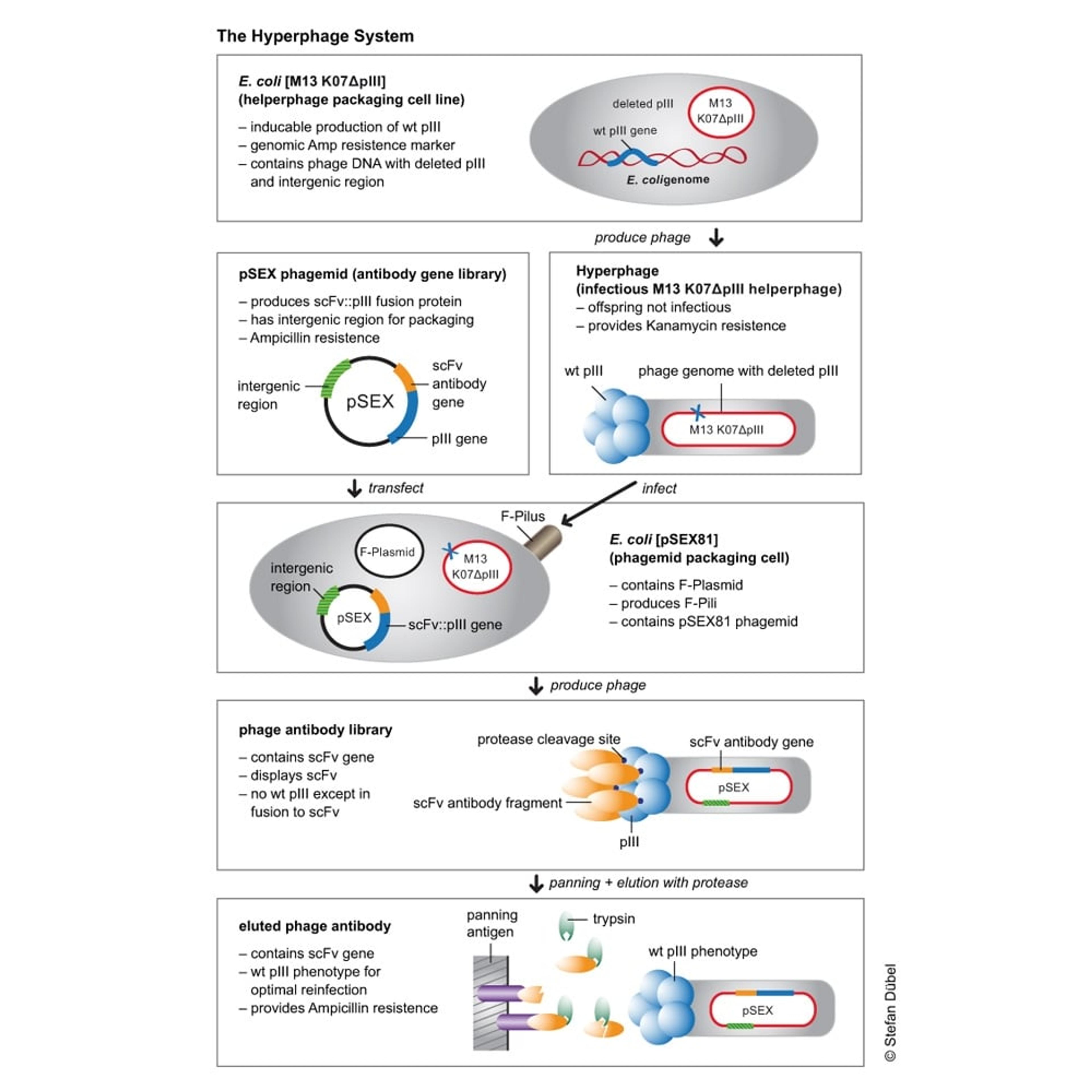
The Hyperphage System
Hyperphages carry a deletion in the pIII gene. They are generated by an E. coli packaging cell line producing functional pIII which is used to package a phage genome with a pIII deletion. The resulting hyperphages carry functional pIII on their surface but lack the pIII gene in their genome. These hyperphages can then be used to infect bacteria with a phagemid library. Each of the resulting display phages carries several copies of the antibody or peptide on its surface, thus dramatically increasing panning efficiency.
Example with pSEX Phagemid Antibody Gene Library
Antigen binding was enhanced by more than two orders of magnitude by using hyperphage. Further, since the antibody carrying plasmid (phagemid) encodes a protease cleavage site between pIII and scFv fragment, the hyperphage-packed library can be eluted by protease treatment, allowing to elute the highest affinity binders, plus restoring wild-type infectivity phenotype to optimize the recovery of the antibody gene of interest.
References
Original papers on successful use of hyperphage
Zantow J et al. (2016). Sci. Rep. 6:34337, DOI:10.1038/srep34337
Connor DO et al. (2016). PLoS One. Feb 9;11(2):e0148986. doi: 10.1371/journal.pone.0148986 114.
Derman Y et al. (2016). Toxins 8:25
Rohrbeck A et al. (2016). Toxins 8:100
Miethe S et al. (2016). PLoS ONE 11:e0161446
Blokzijl A. et al. (2016). Mol Cell Proteomics 15:1848-1856
Kibat J et al. (2016). N. Biotech. 33:574-581
Rasetti-Escargueil C et al. (2015). mAbs 6:1161-1177
Miethe S et al. (2015). PLoS ONE 10:e0139905
Becker M et al. (2015). BMC Biotechnology 15:43
Avril A et al. (2015). BMC Biotechnology 15:86
Droste P et al. (2015).BMC Biotechnol.15:57
Zehner M et al. (2015). Immunity 42:850-863
Kügler J et al. (2015). BMC Biotechnol. 15:10
Fuchs M et al. (2014). BMC Biotechnol. 14:68
Hülseweh B et al. (2014). mAbs 6:717-726
Trott M et al. (2014). PLoS ONE 9:e97478
Miethe S et al. (2014). STO Scientific Publications, STO-MP-HFM239-16
Miethe S et al. (2014). mAbs 6:446-459
Schirrmann T et al. (2014). mAbs, J6(2)
Steinwand M et al. (2013). mAbs, 6:204-218
Zhou M et al. (2013). Eur J immunol, 43:499-509
Meyer T et al. (2012). BMC Biotechnol, 12:29
Wezler X et al. (2012). Hum. Antibod. 21:13-28
Rülker T et al. (2012). PLoS ONE, 7(5):e37242. doi:10.1371/journal.pone.0037242
Zhang C. et al. (2012). PLoS ONE 7(1): e30684. doi:10.1371/journal.pone.0030684
Protocols
Hust M., Frenzel, A., Tomszak, F, Kügler, J & Dübel, S. (2014) Antibody Phage Display. In: Dübel, S. & Reichert, J.M. (eds.) Handbook of Therapeutic Antibodies, 2nd ed. Wiley-VCH, Weinheim, ISBN 978-3-527-32937-3 p. 43-76
Hust, M., Frenzel, A., Schirmann, T. and Dübel, S. (2014) Selection of recombinant antibodies from antibody gene libraries. Methods Mol Biol. 1101, 305-320.2
Hust, M., Frenzel, A., Meyer, T., Schirmann, T. and Dübel, S. (2012). Construction of human naive antibody gene libraries. In: Antibody Engineering: Methods and Protocols, Ed: Chames, P., Meth Mol Biol 907, 85-107
Breitling, F., Broders, O., Helmsing, S., Hust, M & Dübel, S (2010) Improving phage display throughput by using Hyperphage, miniaturised titration and pVIII (g8p) ELISA. In: Antibody Engineering (2nd ed) Springer Protocols. Vol. 1, ISBN 978-3-642-01143-6, p. 197-206
Broders, O., Breitling, F., and Dübel, S. (2003). Hyperphage - Improving Antibody Presentation in Phage Display. Methods Mol Biol 205, 295-302.