KAPA RNA Library Preparation Kits for Illumina
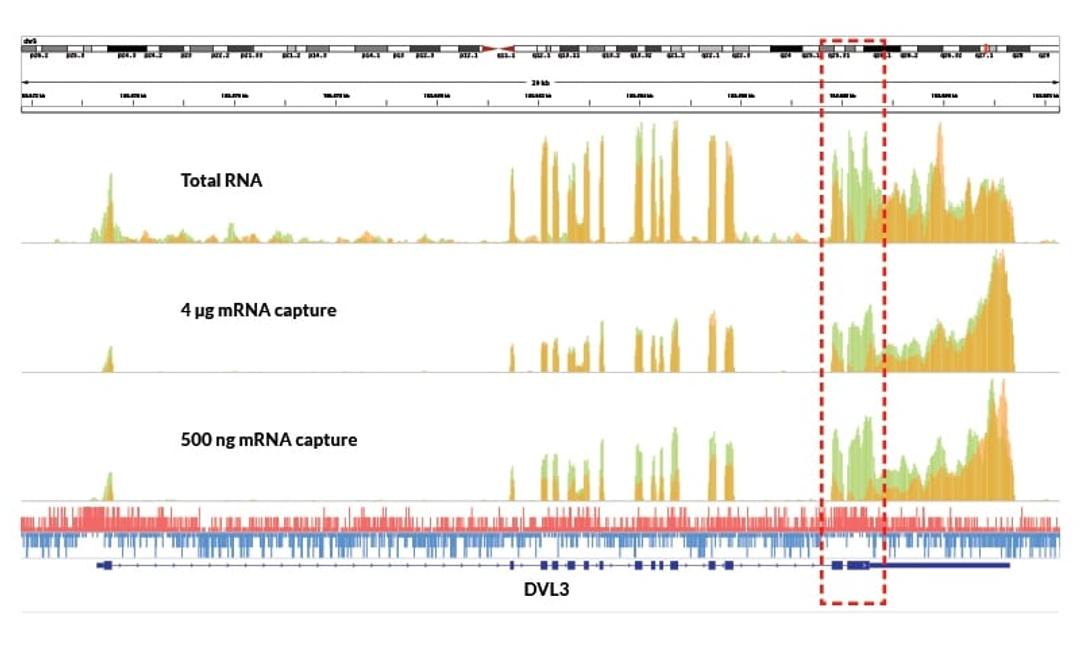
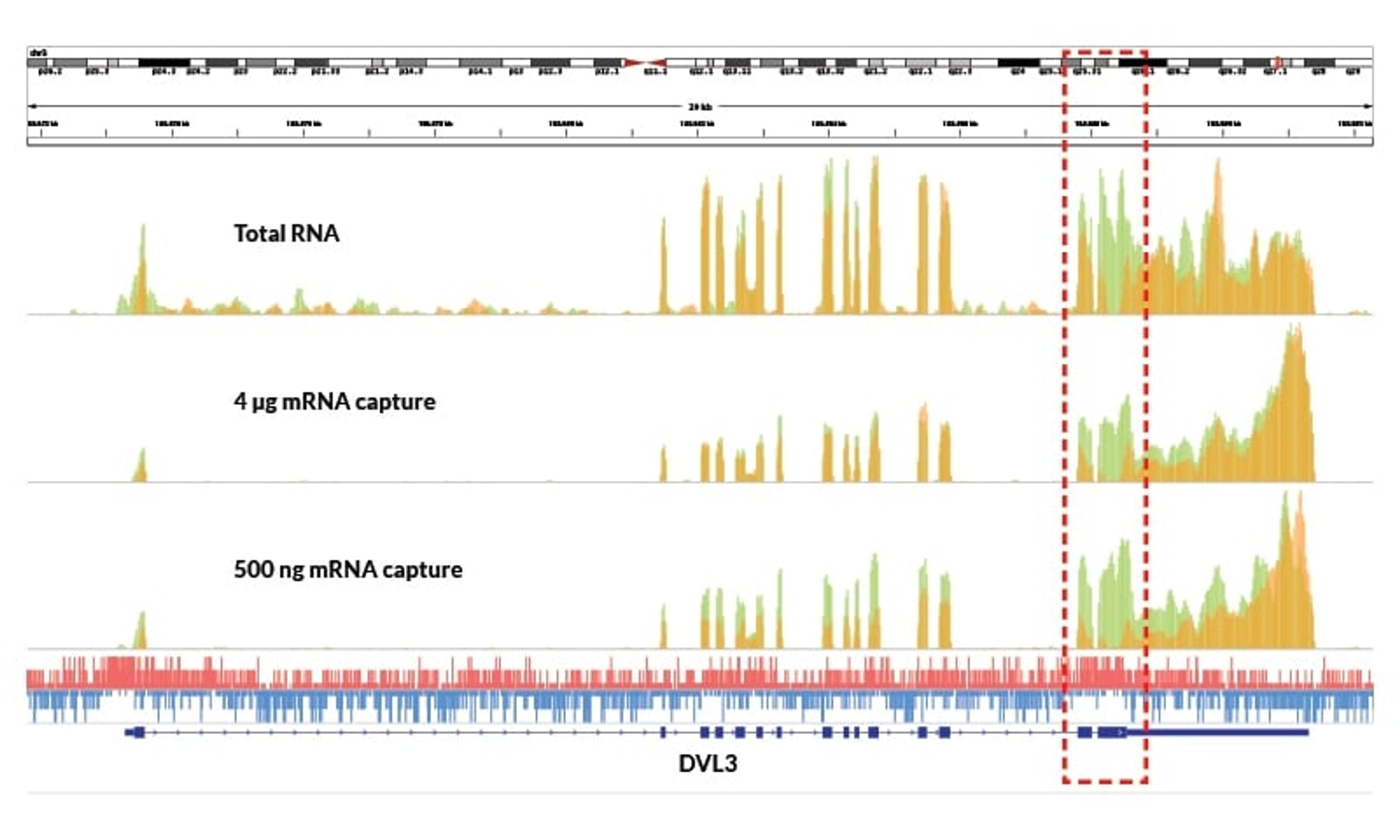
KAPA Stranded mRNA-Seq Kits includes all the enzymes and buffers required for cDNA library preparation for Illumina Next-Generation Sequencing, utilizing 100 ng – 4 µg of total RNA. KAPA mRNA Capture Beads are included for isolation of poly(A)-tailed RNA. Kits provides precise measurement of strand orientation (>99%), uniform coverage, and high-confidence mapping of alternate transcripts, and are optimized for the improved…

The supplier does not provide quotations for this product through SelectScience. You can search for similar products in our Product Directory.
KAPA Stranded mRNA-Seq Kits includes all the enzymes and buffers required for cDNA library preparation for Illumina Next-Generation Sequencing, utilizing 100 ng – 4 µg of total RNA.
KAPA mRNA Capture Beads are included for isolation of poly(A)-tailed RNA. Kits provides precise measurement of strand orientation (>99%), uniform coverage, and high-confidence mapping of alternate transcripts, and are optimized for the improved coverage of GC-rich and low-abundance transcripts. Kits contain KAPA HiFi for high-efficiency and low bias library amplification, as well as KAPA mRNA Capture Beads and a streamlined, “with-bead” protocol.
KAPA Stranded RNA-Seq Kits include all the enzymes and buffers required for cDNA library preparation for Illumina Next-Generation Sequencing, but do not contain the KAPA mRNA Capture Beads. Kits can be used to prepare libraries from 10-400 ng of either poly(A)-selected, ribosomally-depleted, or total RNA.
Features:
Uncover challenging transcripts
- Improved coverage of GC-rich transcripts
- Enhanced identification of exonic regions
Detect low-abundance transcripts
- Enables identification of transcripts missed by competitor kits, even with high input
- High uniformity across varying amounts of sample input
Identify more genes
- Higher percentage of uniquely mapped reads compared to Illumina TruSeq™ Stranded mRNA Sample Prep Kits
- Lower duplication rates yield better coverage
Maintain high coverage uniformity
- Minimal 5′–3′ bias across transcripts
- More uniform distribution of reads over each transcript
Applications:
- Gene expression
- Single nucleotide variation (SNV) discovery
- Post-transcriptional SNVs
- Fusion gene identification
- Targeted transcriptome
- Whole transcriptome